Cover images provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
See the 2020 paper titled "DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL" in Nucleic Acids Research and the corresponding Supplemental PDF for details. Many thanks to Drs. Wilma Olson and Cathy Lawson for their help in the preparation of the illustrations.
Details on how to reproduce the cover images are available on the 3DNA Forum.

Structure of the human minor spliceosome pre-B complex (PDB id: 8Y7E; Bai R, Yuan M, Zhang P, Luo T, Shi Y, Wan R. 2024. Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome. Science 383: 1245–1252). The protein–RNA assembly reveals the mechanisms of recognition and recruitment of several small nuclear ribonucleoproteins (snRNPs) involved in the splicing of U12-type introns. The pre-mRNA is depicted by a red ribbon, and the U12 small nuclear RNA (snRNA) by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are shown as gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Human tRNA splicing endonuclease (TSEN) complex bound to pre-tRNAArg (PDB id: 7UXA; Hayne CK, Butay KJ, Stewart ZD, Krahn JM, Perera L, Williams JG, Petrovitch RM, Deterding LJ, Matera AG, Borgnia MJ, Stanley RE. 2023. Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex. Nat Struct Mol Biol 30: 824–833). Cryo-EM structure of the TSEN protein assembly with pre-tRNAArg provides insights into the recognition and splicing of an intron that must be removed from the pre-tRNA before translation. The pre-tRNAArg is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the TSEN subunits are shown as gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Systemic RNA interference defective protein 1 (SID1) in complex with dsRNA (PDB id: 8XC1; Wang R, Cong Y, Qian D, Yan C, Gong D. 2024. Structural basis for double-stranded RNA recognition by SID1. Nucleic Acids Res 52: 6718–6727). The cryo-EM structure provides a major step towards understanding the mechanism of dsRNA recognition by SID1, involving extensive interactions between basic amino-acid residues and the sugar-phosphate backbone. The dsRNA chains are depicted by red, green, blue, and yellow ribbons, with bases and Watson-Crick base pairs represented as color-coded blocks and minor-groove edges colored white: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; SID1 is shown by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Complex of arginyl-tRNA-protein transferase 1 (ATE1) with tRNAArg and a short peptide substrate (PDB id: 8UAU; Lan X, Huang W, Kim SB, Fu D, Abeywansha T, Lou J, Balamurugan U, Kwon YT, Ji CH, Taylor DJ, Zhang Y. 2024. Oligomerization and a distinct tRNA-binding loop are important regulators of human arginyl-transferase function. Nat Commun 15: 6350). The ATE1 homodimer dissociates upon binding the peptide and forms a loop that wraps around tRNAArg. The tRNAArg is depicted by a red ribbon, with bases and Watson–Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; ATE1 is shown by a gold ribbon and the peptide by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of endoribonuclease P (RNase P) in complex with pre-tRNAHis-Ser (PDB id: 8CBK; Meynier V, Hardwick SW, Catala M, Roske JJ, Oerum S, Chirgadze DY, Barraud P, Yue WW, Luisi BF, Tisné C. 2024. Structural basis for human mitochondrial tRNA maturation. Nat Commun 15: 4683). The structure reveals the first step of human mitochondrial tRNA maturation by RNase P, processing the 5′-leader of pre-tRNA. The RNA is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the protein assembly is shown by the gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of a group II intron ribonucleoprotein in the pre-ligation state (PDB id: 8T2R; Xu L, Liu T, Chung K, Pyle AM. 2023. Structural insights into intron catalysis and dynamics during splicing. Nature 624: 682–688). The pre-ligation complex of the Agathobacter rectalis group II intron reverse transcriptase/maturase with intron and 5′-exon RNAs makes it possible to construct a picture of the splicing active site. The intron is depicted by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the 5′-exon is shown by white spheres and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Complex of terminal uridylyltransferase 7 (TUT7) with pre-miRNA and Lin28A (PDB id: 8OPT; Yi G, Ye M, Carrique L, El-Sagheer A, Brown T, Norbury CJ, Zhang P, Gilbert RJ. 2024. Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs. Nat Struct Mol Biol 31: 1426–1438). The RNA-binding pluripotency factor LIN28A invades and melts the RNA and affects the mechanism of action of the TUT7 enzyme. The RNA backbone is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; TUT7 is represented by a gold ribbon and LIN28A by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Cryo-EM structure of the pre-B complex (PDB id: 8QP8; Zhang Z, Kumar V, Dybkov O, Will CL, Zhong J, Ludwig SE, Urlaub H, Kastner B, Stark H, Lührmann R. 2024. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 630: 1012–1019). The pre-B complex is thought to be critical in the regulation of splicing reactions. Its structure suggests how the cross-exon and cross-intron spliceosome assembly pathways converge. The U4, U5, and U6 snRNA backbones are depicted respectively by blue, green, and red ribbons, with bases and Watson-Crick base pairs shown as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are represented by gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the Hendra henipavirus (HeV) nucleoprotein (N) protein-RNA double-ring assembly (PDB id: 8C4H; Passchier TC, White JB, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN. 2024. The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Sci Rep 14: 14099). The HeV N protein adopts a bi-lobed fold, where the N- and C-terminal globular domains are bisected by an RNA binding cleft. Neighboring N proteins assemble laterally and completely encapsidate the viral genomic and antigenomic RNAs. The two RNAs are depicted by green and red ribbons. The U bases of the poly(U) model are shown as cyan blocks. Proteins are represented as semitransparent gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the helicase and C-terminal domains of Dicer-related helicase-1 (DRH-1) bound to dsRNA (PDB id: 8T5S; Consalvo CD, Aderounmu AM, Donelick HM, Aruscavage PJ, Eckert DM, Shen PS, Bass BL. 2024. Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA. eLife 13: RP93979. Cryo-EM structures of Dicer-1 in complex with DRH-1, RNAi deficient-4 (RDE-4), and dsRNA provide mechanistic insights into how these three proteins cooperate in antiviral defense. The dsRNA backbone is depicted by green and red ribbons. The U-A pairs of the poly(A)·poly(U) model are shown as long rectangular cyan blocks, with minor-groove edges colored white. The ADP ligand is represented by a red block and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Moreover, the following 30 [12(2021) + 12(2022) + 6(2023)] cover images of the RNA Journal were generated by the NAKB (nakb.org).
Cover image provided by the Nucleic Acid Database (NDB)/Nucleic Acid Knowledgebase (NAKB; nakb.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Recently, I came across the so-called Calcutta U-U base pair (bp) [see figure below] while reading articles on C-H…O contacts in nucleic acid structures. Not familiar with this named pair before, I was curious to find out what it’s about. After some searching, I traced the origin of the Calcutta U-U bp to the following two papers published by Sundaralingam’s group during the middle 1990s:
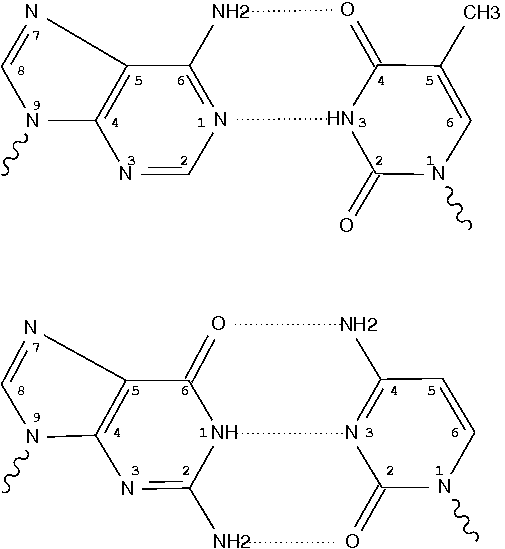
We have called the novel U•U base pair, where the Hoogsteen face of one of the pyrimidines is involved in a C5-H—O4 hydrogen bond, the ‘Calcutta Base Pair’, since it was announced at the International Seminar-cum-School on Macromolecular Crystallographic Data held in Calcutta, November 16-20, 1995.
We recently discovered a novel U•U base pair, referred to as the Calcutta base pair, in the crystal structure of an RNA hexamer UUCGCG (Ref. 18). The two uracil bases form a conventional N(3)-H…O(4) and an unconventional C(5)-H…O(2) hydrogen bond (Fig. 3a). The C-H…O interaction is entirely ‘voluntary’ and not ‘forced’, underlining its importance in base mispairing.
3DNA has no problem to identify the Calcutta U-U bps (or any pair for that matter); an example is shown below based on the RNA hexamer UUCGCG structure (PDB entry: 1osu) solved by Sundaralingam and colleagues.
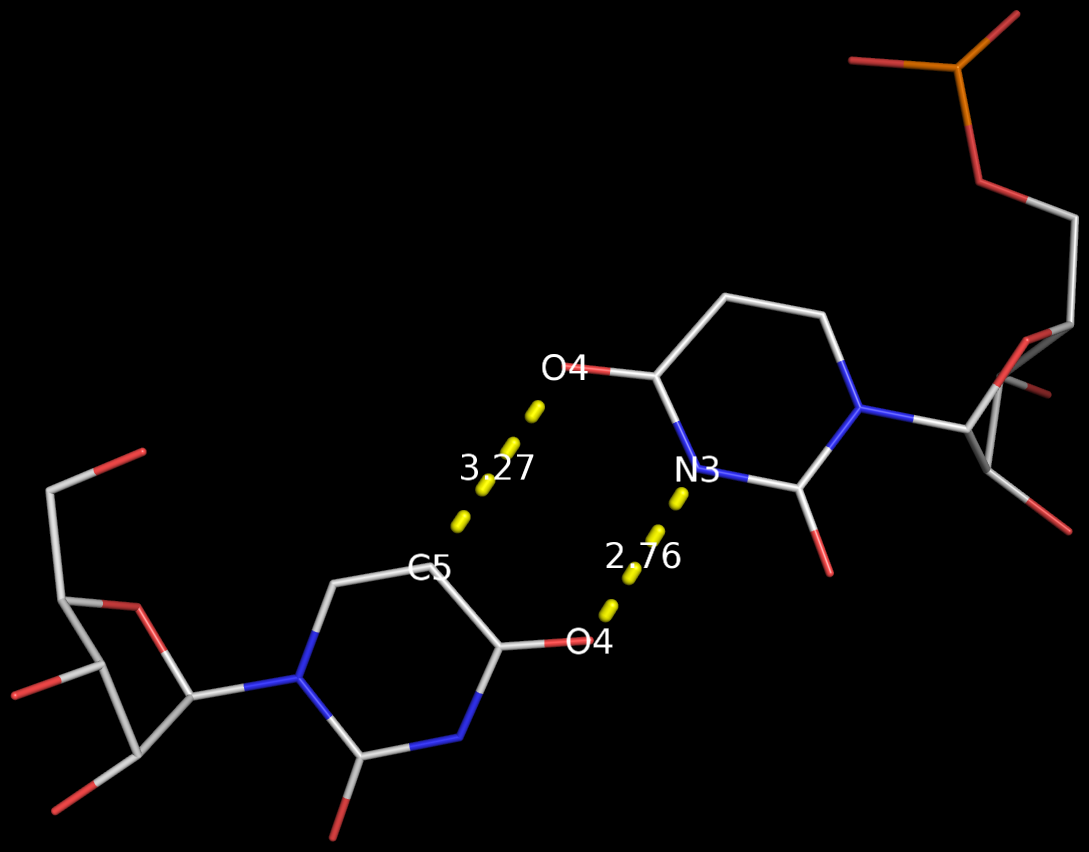
In the new 3DNA component I’ve been working on (and to be released soon), the Calcutta U-U pair is characterized as below:
1/A.U1 3/A.U2 [U-U] Calcutta 00-n/a tHW -MW
anti C3'-endo 8.9 --- anti C3'-endo 30.3
dcc=11.18 dnn=8.48 dmm=7.58 tor=-174.1
H-bonds[2]: "O4(carbonyl)-N3(imino)[2.76]; C5-O4(carbonyl)[3.27]"
Shear=-3.67 Stretch=-0.52 Stagger=-0.89
Buckle=-1.41 Propeller=-16.03 Opening=-90.67
The Calcutta pair is explicitly named, along with other named base pairs (e.g., Watson-Crick [WC], Wobble, and Hoogsteen bps). It is classified as type tHW (trans with Hoogsteen/WC interacting edges), following the commonly used Leontis-Westhof nomenclature. It does not belong to any of the 28 bps (00-n/a) with at least two conventional H-bonds, as categorized by Saenger. In 3DNA, the Calcutta U-U pair is of M-N type, designated as -MW.
Among the well-known named base pairs, some are after the scientists who discovered them (e.g., WC and Hoogsteen bps), while others are based on chemical/geometrical features (e.g., Wobble and Sheared G-A bps), or a combination of both (e.g., reversed WC/Hoogsteen bps). The Calcutta U-U pair is unique in that it is named after a place in India:
Kolkata, or Calcutta, is the capital of the Indian state of West Bengal. … While the city’s name has always been pronounced Kolkata or Kolikata in Bengali, the anglicized form Calcutta was the official name until 2001, when it was changed to Kolkata in order to match Bengali pronunciation.

Prior to v2.1, 3DNA does not provide any direct support for the analysis of molecular dynamics (MD) simulations trajectories of nucleic acid structures. Nevertheless, over the years, I noticed some significant applications of 3DNA in the active MD field; see my blog post (December 6, 2009) titled 3DNA in the PCCP nucleic acid simulations themed issue. In January 2011, I released a set of two Ruby scripts specifically aimed to facilitate the analysis of MD simulations trajectories. Thereafter (as of 3DNA v2.1), I have significantly refined and expanded the Ruby scripts, and consolidated the functionality under one umbrella, x3dna_ensemble with multiple sub-commands (analyze, block_image, extract, and reorient). I believe x3dna_ensemble would make it straightforward to analyze ensembles (NMR or MD simulations trajectories) of nucleic acid structures.
Under this background, I am glad to read recently an article titled Structure, Stiffness and Substates of the Dickerson-Drew Dodecamer in J. Chem. Theory Comput. where 3DNA was used extensively. This work represents a re-visit of the classic Dickerson−Drew B-DNA dodecamer d-[CGCGAATTCGCG]2 using state-of-the-art MD simulations with different ionic conditions and solvation models, and compares the MD trajectories with modern crystallographic and NMR data. Among the author list (Tomas Drsata, Alberto Perez, Modesto Orozco, Alexandre Morozov, Jiri Sponer, and Filip Lankas) are some well-known figures in the MD field of nucleic acid structures.
Reading through the text, I am not sure if the newly available functionality of x3dna_ensemble was used. From the excerpts of the citations given below, however, it seems obvious that 3DNA is now well-accepted by the MD community.
Snapshots taken in 10 ps intervals were analyzed using the 3DNA program.43 From 3DNA outputs, time series of conformational parameters were extracted. These included the intra-base-pair coordinates (buckle, propeller, opening, shear, stretch, and stagger), inter-base-pair or step coordinates (tilt, roll, twist, shift, slide, and rise) as well as groove widths (based on P−P distances), backbone torsions, and sugar puckers.
Contrary to the original work of Lankas et al.,31 the intra-base-pair and step coordinates used here are those defined by 3DNA.43
Here, we apply this model together with the 3DNA definitions of the intra-base-pair and step coordinates.43
However, important differences remain, and non- negligible differences are in fact observed between individual experimental structures also in the central part of DD, even though the intra-base-pair and step coordinates are computed using the same coordinate definitions64 (we consistently use the 3DNA coordinates in this work).

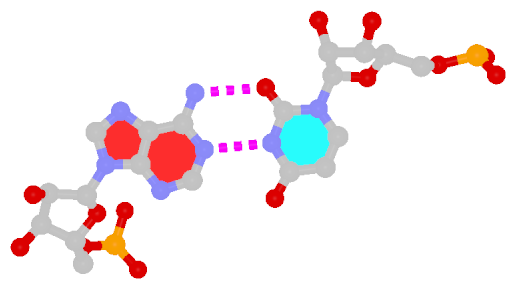
In the field of nucleic acid structures, especially in the ‘RNA world’, we often hear named base pairs (bp). Among those, the Watson-Crick (WC) A–U and G–C bps (see figure below) are by far the most common.
Closely related to the WC bps are the so-called reversed WC (rWC) bps, where the relative glycosidic bond are reversed; instead of being on the same side of the bases as in WC bps shown above, they are now on opposite sides in rWC bps as shown below. According to the Leontis-Westhof (LW) bp classification scheme, the rWC bps belong to trans WC/WC. Following Saenger’s numbering, the rWC A+U bp corresponds to XXI, and the rWC G+C bp XXII.
In the figures below, the name of each type of bp and its LW & Saenger designations (separated by ‘;’) are noted under the corresponding image. All images are generated with 3DNA; for easy comparison, each bp is oriented in the reference frame of the leading base.
 |
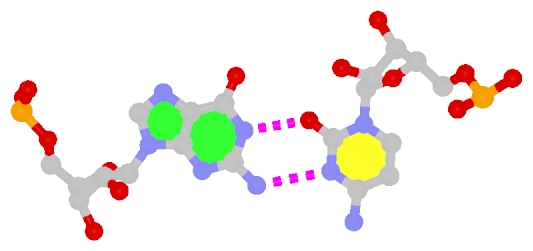 |
| Reversed WC A+U pair |
Reversed WC G+C pair |
| trans WC/WC; XXI |
trans WC/WC; XXII |
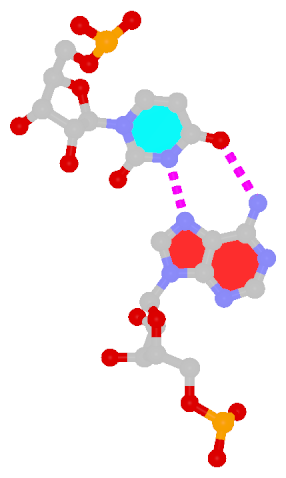
The next most famous one is the Hoogsteen A+U bp, which also has a reverse variant, i.e., the rHoogsteen A–U bp (see figure below). Now the major groove edge of A, termed the Hoogsteen edge by LW, is used for pairing with U.
 |
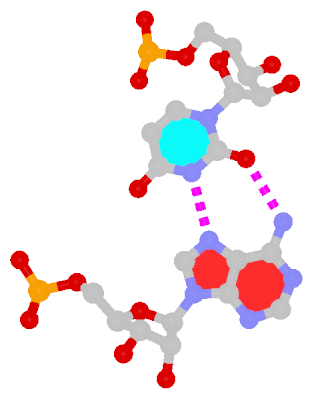 |
| Hoogsteen A+U pair |
Reversed Hoogsteen A–U pair |
| cis Hoogsteen/WC; XXIII |
trans Hoogsteen/WC; XXIV |
First proposed by Crick in 1966 to account for the degeneracy in codon–anticodon pairing, the Wobble bp is an essential component (in addition to the WC bps) in forming double helical RNA secondary structures.
 |
| Wobble G–U pair |
| cis WC/WC; XXVIII |
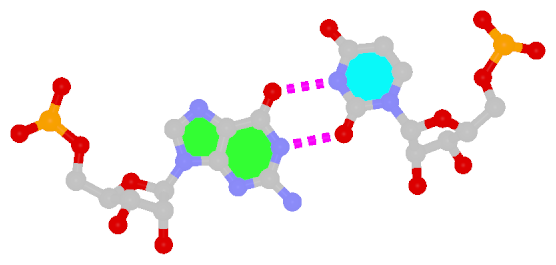
The sheared G–A base pair
Sheared G–A is a commonly found non-WC bp in both DNA and RNA structures. Noticeably, tandem sheared G–A bps introduce distinct stacking geometry. Here G uses its minor groove edge, termed the sugar edge by LW, to pair with the Hoogsteen edge of A.
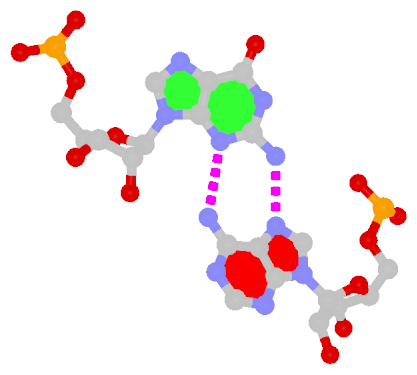 |
| Sheared G–A pair |
| trans Suger/Hoogsteen; XI |
Dinucleotide platforms
Dinucleotide platforms are formed via side-by-side pairing of adjacent bases; the most common of which are GpU and ApA. Here the sugar (minor-groove) edge of the 5′ base interacts with the Hoogsteen (major-groove) edge of the 3′ base. Since there is only one base-base H-bond in dinucleotide platforms, no Saenger classification is available. In 3DNA output, the GpU dinucleotide platform is designated as G+U, and ApA as A+A.
 |
 |
| GpU dinucleotide platform |
ApA dinucleotide platform |
| cis Sugar/Hoogsteen; n/a |
cis Sugar/Hoogsteen; n/a |
Other named base pairs
There exist other named bps in RNA literature, e.g., G⋅A imino, A⋅C reverse Hoogsteen, G⋅U reverse Wobble etc. In the my experience, they are (much) less commonly used than the ones illustrated above.

Glycosidic bond “is a type of covalent bond that joins a carbohydrate (sugar) molecule to another group, which may or may not be another carbohydrate.” In nucleic acid structures, the other group is a nucleobase, and the predominated type is the N-glycosidic bond where the purine (A/G) N9 or pyrimidine (C/T/U) N1 atom connects to the C1′ atom of the five-membered (deoxy) ribose sugar ring. Another well-known type is the C-glycosidic bond in pseudouridine, the most common modified base in RNA structures where the C5 atom instead of N1 is linked to the C1′ atom of the sugar ring.

Recently, I performed a survey of all nucleic-acid-containing structures in the PDB/NDB database to see how many types of glycosidic bond are there. As always, I noticed some inconsistencies in the data: nucleotides with disconnected base/sugar, a base labeled as U but with pseudoU-type C-glycosidic bond. Shown below are a few unusual types of glycosidic bond in otherwise seemingly “normal” structures:
- The residue GN7 (number 28 on chain A) in PDB entry 1gn7 contains a N7-glycosylated guanine.
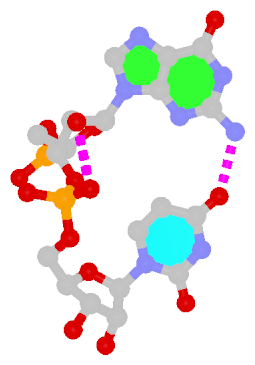
- The residue UPG (number 501 on chain A) in PDB entry 1y6f has sugar C1C (instead of C1′) atom connects to N1 of U.
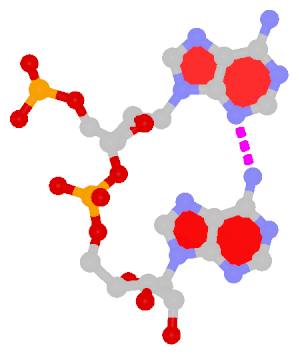
- The residue XAE (number 11 on chain B) in PDB entry 2icz contains a benzo-homologous adenine.
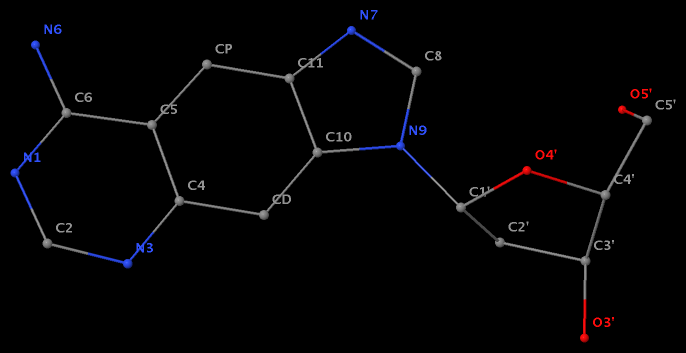
- The residue F5H (number 206 on chain B) in PDB entry 3v06 has N1 of U connects to C2′ of a six-membered sugar ring.
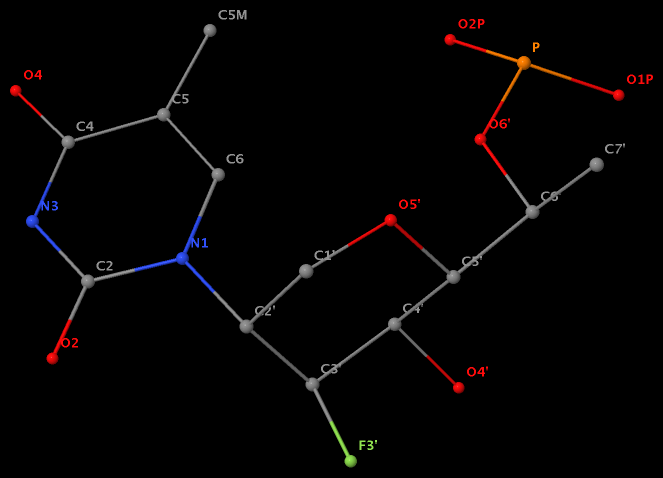
The unusual glycosidic bond has implications in 3DNA calculated parameters, for example the chi torsion angle. Identifying such cases would help refine 3DNA to provide sensible parameters and to avoid possible misinterpretations.

As of today (2012-09-16), the number of 3DNA forum registrations has reached 500! A quick browse of the ‘Statistics Center’ shows that over 80% of the registrations (400+) are after March 2012, when the new 3DNA homepage/forum were launched.
The sharp increase in registration is mostly due to the streamlined, web-based way to distribute the 3DNA software package. As far as I know, the number of 3DNA registrations/downloads in the past six months is significantly higher than that of 3DNA v2.0 for over three years. Equally importantly, I have been able to fixed every reported bug, addressed each feature request, and updated the 3DNA v2.1 distribution promptly.
I also feel confident to declare that up to now, the 3DNA Forum is spam free (at least to the extent I am aware). To this end, I’ve taken the following three measures:
- Installation of the SMF “Mod Stop Spammer”; as of this writing, it shows “3920 Spammers blocked up until today”.
- By using 3DNA-related verification questions. At its current setting, a user must answer correctly three of the ‘simple’ yet effective verification questions. Early on, I decided deliberately not to use CAPTCHA as an anti-spam means, based on my past experience.
- I’ve continuously monitored (new) registrations, and taken immediate actions against any suspicious registration. Due to the effectiveness of above two steps, so far I only have to manually handle just a few spam registrations. Nevertheless, it does illustrate the fact that no automatic method is perfect, and expert inspection is required to ensure desired results.
Overall, the new simplified way to distribute the 3DNA software package is working as intended; now users can easily access all distributed versions of 3DNA, and I can focus on support and further development of the software.

From v1.5 or even earlier on, 3DNA provides an automatic classification of a dinucleotide step into A-, B- or TA-DNA conformation. Figure 5 of the 2003 3DNA Nucleic Acids Research paper (NAR03) shows three sets of scatter plots — helical inclination and x‐displacement, dimer step Roll and Slide, and the projected phosphorus z coordinates Zp and Zp(h) — to differentiate the A-, B- and TA-DNA dinucleotide steps.

Among the criteria tested, the most discriminative ones are the projected phosphorus z coordinates, Zp in the middle step frame (see figure below), and Zp(h) defined similarly but in the middle helical frame.
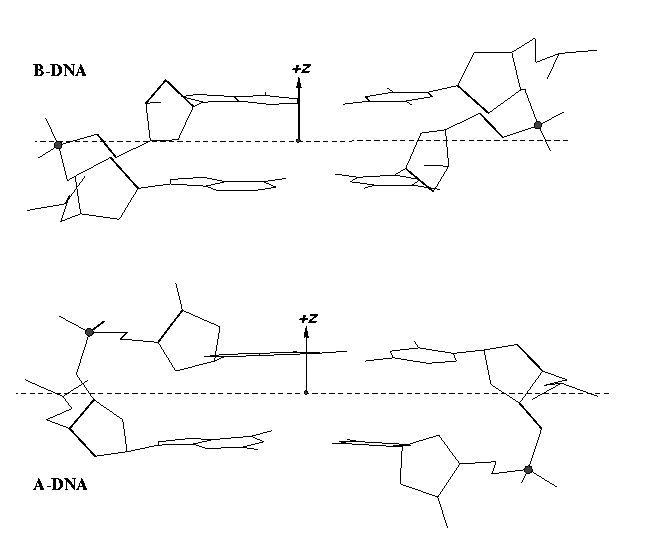
Over the years, I have received many questions regarding the datasets used in generating Figure 5 of NAR03. Back in August 2006, a user asked for IDs of the TA-DNA structures — see DNA standards/statistics using 3DNA. In April 2007, another user requested the same TA-DNA dataset. Early this year, a user asked for 3DNA’s A-DNA definition. More recently, yet another user would like to ask about the DNA set used for the analysis that is presented in Fig 5. in the NAR 2003 paper.
I am glad to see that after nearly a decade of the NAR03 publication, the user community is still interested in knowing details in the work. So I decided to dig into my archive for the original data files and scripts used to generate Figure 5 of NAR03. It was not an easy journey; just releasing the data files and scripts is not enough, I’d like to verify that they work together as intended in today’s computing environment. Luckily, I am finally able to get to the bottom of the issues. The details are in the post Datasets and scripts for reproducing Figure 5 of the 3DNA NAR03 paper. The tarball file named 3DNA-NAR03-Fig5.tar.gz is available by clicking the link.

As noted in post Rectangular block expressed in MDL molfile format, I added the -mol option (in v2.1) to convert 3DNA’s native alchemy to the better-supported MDL molfile format, to make the characteristic schematic representations more widely accessible. Along the line, I have recently further augmented alc2img with the -pdb option to transform alchemy to the PDB format.
While the macromolecular PDB format is certainly not convenient for specifying linkage details of small molecules, it’s nevertheless the best-documented and by far the most widely supported than molfile or alchemy in currently available molecular viewers. For example, the PDB format is consistently supported in Jmol, PyMOL, RasMol, DeepView, and UCSF Chimera. Moreover, the PDB format does have the CONECT section to provide information on atomic connectivity:
The CONECT records specify connectivity between atoms for which coordinates are supplied. The connectivity is described using the atom serial number as shown in the entry. CONECT records are mandatory for HET groups (excluding water) and for other bonds not specified in the standard residue connectivity table.
The alc2img -pdb option takes advantage of the CONECT records and specifies all ‘bond’ linkages explicitly. The usage is very simple — take the standard base-pair rectangular block file (‘Block_BP.alc’) as an example, the conversion can be performed as below:
alc2img -pdb Block_BP.alc Block_BP.pdb
Content of ‘Block_BP.alc’
12 ATOMS, 12 BONDS
1 N -2.2500 5.0000 0.2500
2 N -2.2500 -5.0000 0.2500
3 N -2.2500 -5.0000 -0.2500
4 N -2.2500 5.0000 -0.2500
5 C 2.2500 5.0000 0.2500
6 C 2.2500 -5.0000 0.2500
7 C 2.2500 -5.0000 -0.2500
8 C 2.2500 5.0000 -0.2500
9 C -2.2500 5.0000 0.2500
10 C -2.2500 -5.0000 0.2500
11 C -2.2500 -5.0000 -0.2500
12 C -2.2500 5.0000 -0.2500
1 1 2
2 2 3
3 3 4
4 4 1
5 5 6
6 6 7
7 7 8
8 5 8
9 9 5
10 10 6
11 11 7
12 12 8
Content of ‘Block_BP.pdb’
REMARK 3DNA v2.1 (c) 2012 Dr. Xiang-Jun Lu (http://home.x3dna.org)
HETATM 1 N ALC A 1 -2.250 5.000 0.250 1.00 1.00 N
HETATM 2 N ALC A 1 -2.250 -5.000 0.250 1.00 1.00 N
HETATM 3 N ALC A 1 -2.250 -5.000 -0.250 1.00 1.00 N
HETATM 4 N ALC A 1 -2.250 5.000 -0.250 1.00 1.00 N
HETATM 5 C ALC A 1 2.250 5.000 0.250 1.00 1.00 C
HETATM 6 C ALC A 1 2.250 -5.000 0.250 1.00 1.00 C
HETATM 7 C ALC A 1 2.250 -5.000 -0.250 1.00 1.00 C
HETATM 8 C ALC A 1 2.250 5.000 -0.250 1.00 1.00 C
HETATM 9 C ALC A 1 -2.250 5.000 0.250 1.00 1.00 C
HETATM 10 C ALC A 1 -2.250 -5.000 0.250 1.00 1.00 C
HETATM 11 C ALC A 1 -2.250 -5.000 -0.250 1.00 1.00 C
HETATM 12 C ALC A 1 -2.250 5.000 -0.250 1.00 1.00 C
CONECT 1 2 4
CONECT 2 1 3
CONECT 3 2 4
CONECT 4 1 3
CONECT 5 6 8 9
CONECT 6 5 7 10
CONECT 7 6 8 11
CONECT 8 5 7 12
CONECT 9 5
CONECT 10 6
CONECT 11 7
CONECT 12 8
END

From a pure structural perspective, the designation of the two strands in an anti-parallel DNA duplex is sort of arbitrary. Thus, for a given PDB file, let’s assume that the atomic coordinates of chain A (strand I) come before those of chain B (strand II). We can swap the order of the two chains as they appear in the PDB file, i.e., list first the atomic coordinates of chain B and then those of chain A.
Structurally, the two settings corresponding to exactly the same DNA molecule. As far as 3DNA goes, however, the different orderings do make a different in calculated parameters. Using the Dickerson B-DNA dodecamer CGCGAATTCGCG solved at high resolution (PDB entry 355d) as an example, running 3DNA find_pair and analyze on ‘355d.pdb’ gives the results (abbreviated) below:
find_pair 355d.pdb 355d.bps
# contents of file '355d.bps':
------------------------------------------------------------------
355d.pdb
355d.out
2 # duplex
12 # number of base-pairs
1 1 # explicit bp numbering/hetero atoms
1 24 0 # 1 | ....>A:...1_:[.DC]C-----G[.DG]:..24_:B<....
2 23 0 # 2 | ....>A:...2_:[.DG]G-----C[.DC]:..23_:B<....
3 22 0 # 3 | ....>A:...3_:[.DC]C-----G[.DG]:..22_:B<....
4 21 0 # 4 | ....>A:...4_:[.DG]G-----C[.DC]:..21_:B<....
5 20 0 # 5 | ....>A:...5_:[.DA]A-----T[.DT]:..20_:B<....
6 19 0 # 6 | ....>A:...6_:[.DA]A-----T[.DT]:..19_:B<....
7 18 0 # 7 | ....>A:...7_:[.DT]T-----A[.DA]:..18_:B<....
8 17 0 # 8 | ....>A:...8_:[.DT]T-----A[.DA]:..17_:B<....
9 16 0 # 9 | ....>A:...9_:[.DC]C-----G[.DG]:..16_:B<....
10 15 0 # 10 | ....>A:..10_:[.DG]G-----C[.DC]:..15_:B<....
11 14 0 # 11 | ....>A:..11_:[.DC]C-----G[.DG]:..14_:B<....
12 13 0 # 12 | ....>A:..12_:[.DG]G-----C[.DC]:..13_:B<....
------------------------------------------------------------------
analyze 355d.bps
# generate output file '355d.out', with base-pair step parameters:
****************************************************************************
step Shift Slide Rise Tilt Roll Twist
1 CG/CG 0.09 0.04 3.20 -3.22 8.52 32.73
2 GC/GC 0.50 0.67 3.69 2.85 -9.06 43.88
3 CG/CG -0.14 0.59 3.00 0.97 11.30 25.11
4 GA/TC -0.45 -0.14 3.39 -1.59 1.37 37.50
5 AA/TT 0.17 -0.33 3.30 -0.33 0.46 37.52
6 AT/AT -0.01 -0.60 3.22 -0.31 -2.67 32.40
7 TT/AA -0.08 -0.40 3.22 1.68 -0.97 33.74
8 TC/GA -0.27 -0.23 3.47 0.68 -1.69 42.14
9 CG/CG 0.70 0.78 3.07 -3.66 4.18 26.58
10 GC/GC -1.31 0.36 3.37 -2.85 -9.37 41.60
11 CG/CG -0.31 0.21 3.17 -0.68 6.69 33.31
****************************************************************************
Reversing the order of chains A and B in ‘355d.pdb’ as ‘355d-reversed.pdb’ and repeating the above procedure, we have the following results:
find_pair 355d-reversed.pdb 355d-reversed.bps
# contents of file '355d-reversed.bps':
------------------------------------------------------------------
355d-reversed.pdb
355d-reversed.out
2 # duplex
12 # number of base-pairs
1 1 # explicit bp numbering/hetero atoms
1 24 0 # 1 | ....>B:..13_:[.DC]C-----G[.DG]:..12_:A<....
2 23 0 # 2 | ....>B:..14_:[.DG]G-----C[.DC]:..11_:A<....
3 22 0 # 3 | ....>B:..15_:[.DC]C-----G[.DG]:..10_:A<....
4 21 0 # 4 | ....>B:..16_:[.DG]G-----C[.DC]:...9_:A<....
5 20 0 # 5 | ....>B:..17_:[.DA]A-----T[.DT]:...8_:A<....
6 19 0 # 6 | ....>B:..18_:[.DA]A-----T[.DT]:...7_:A<....
7 18 0 # 7 | ....>B:..19_:[.DT]T-----A[.DA]:...6_:A<....
8 17 0 # 8 | ....>B:..20_:[.DT]T-----A[.DA]:...5_:A<....
9 16 0 # 9 | ....>B:..21_:[.DC]C-----G[.DG]:...4_:A<....
10 15 0 # 10 | ....>B:..22_:[.DG]G-----C[.DC]:...3_:A<....
11 14 0 # 11 | ....>B:..23_:[.DC]C-----G[.DG]:...2_:A<....
12 13 0 # 12 | ....>B:..24_:[.DG]G-----C[.DC]:...1_:A<....
------------------------------------------------------------------
analyze 355d-reversed.bps
# generate output file '355d-reversed.out', with base-pair step parameters:
****************************************************************************
step Shift Slide Rise Tilt Roll Twist
1 CG/CG 0.31 0.21 3.17 0.68 6.69 33.31
2 GC/GC 1.31 0.36 3.37 2.85 -9.37 41.60
3 CG/CG -0.70 0.78 3.07 3.66 4.18 26.58
4 GA/TC 0.27 -0.23 3.47 -0.68 -1.69 42.14
5 AA/TT 0.08 -0.40 3.22 -1.68 -0.97 33.74
6 AT/AT 0.01 -0.60 3.22 0.31 -2.67 32.40
7 TT/AA -0.17 -0.33 3.30 0.33 0.46 37.52
8 TC/GA 0.45 -0.14 3.39 1.59 1.37 37.50
9 CG/CG 0.14 0.59 3.00 -0.97 11.30 25.11
10 GC/GC -0.50 0.67 3.69 -2.85 -9.06 43.88
11 CG/CG -0.09 0.04 3.20 3.22 8.52 32.73
****************************************************************************
Comparing the base-pair step parameters between ‘355d.out’ and ’355d-reversed.out’, one would notice that while slide/rise/roll/twist simply switch orders, shift/tilt (the x-axis parameters) also flip their signs. On the other hand, the nucleotide serial numbers specifying base pairs (the left two columns) are identical in ‘355d.bps’ and ’355d-reversed.bps’.
Apart from explicitly swapping the two strands in PDB data file, one can simply switch around the nucleotide serial numbers generated with find_pair in order to analyze a DNA duplex based on its complementary sequence instead of the primary one. For example, starting from the same PDB file ‘355d.pdb’, we change ‘355d.bps’ to ’355d-cs.bps’ as below,
------------------------------------------------------------------
355d.pdb
355d-cs.out
2 # duplex
12 # number of base-pairs
1 1 # explicit bp numbering/hetero atoms
13 12
14 11
15 10
16 9
17 8
18 7
19 6
20 5
21 4
22 3
23 2
24 1
------------------------------------------------------------------
Run analyze 355d-cs.bps, one would get exactly the same parameters in output file ’355d-cs.out’ as in ’355d-reversed.out’.

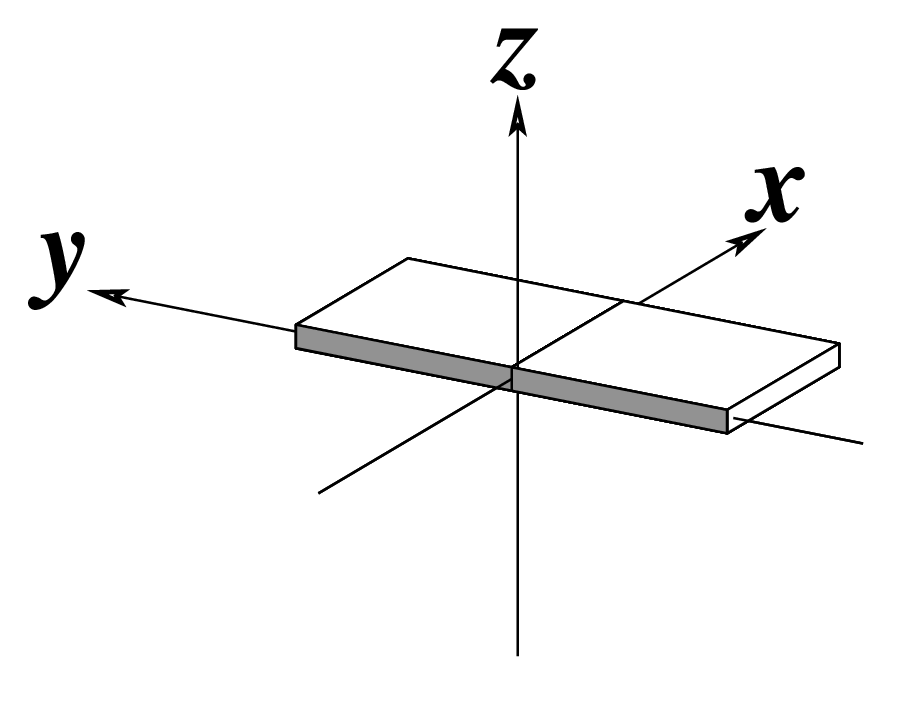
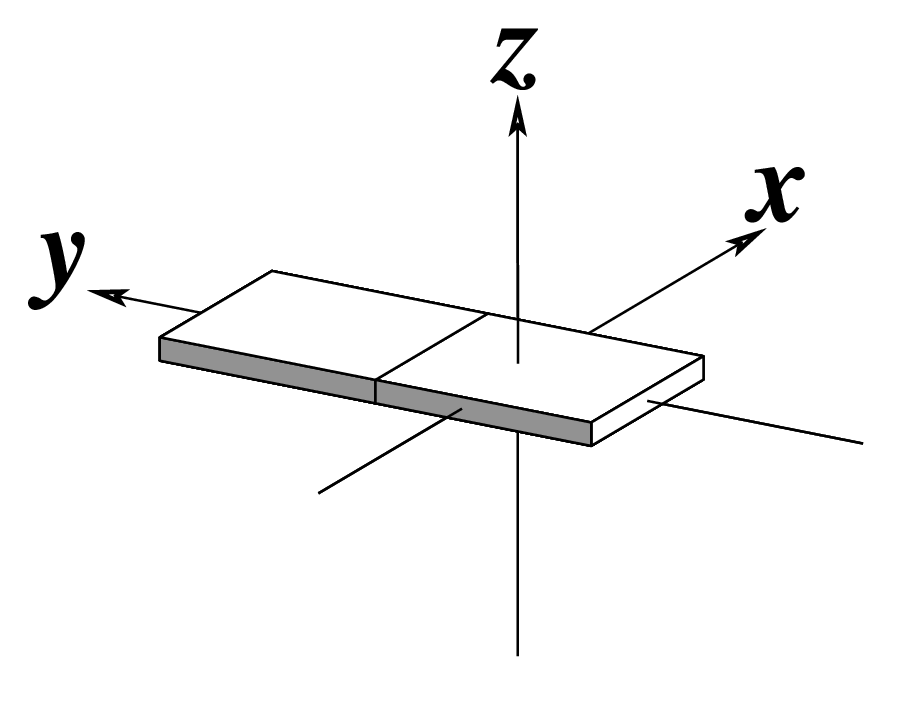
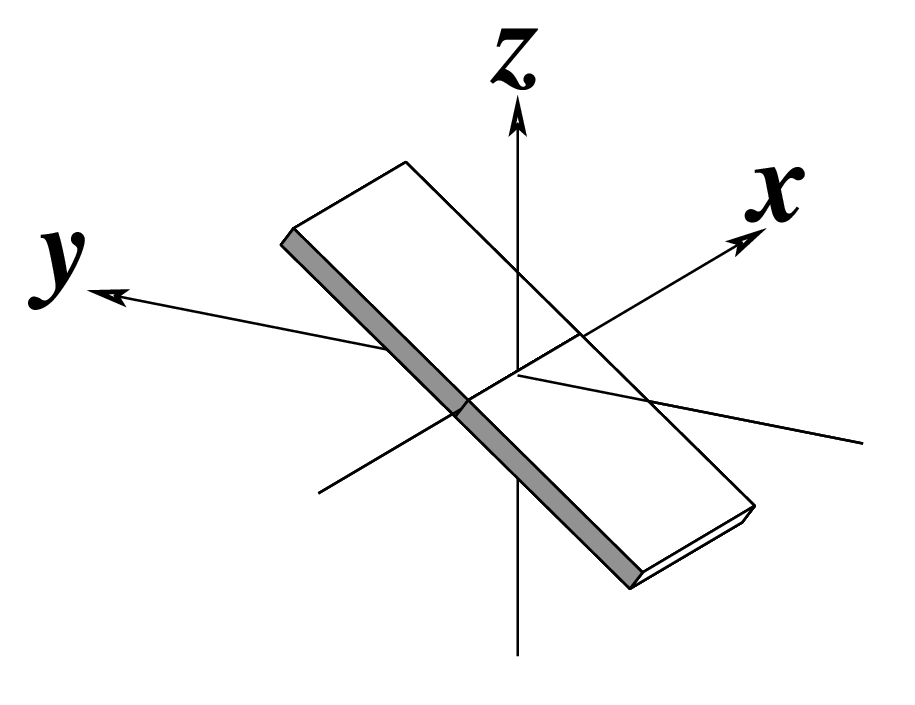
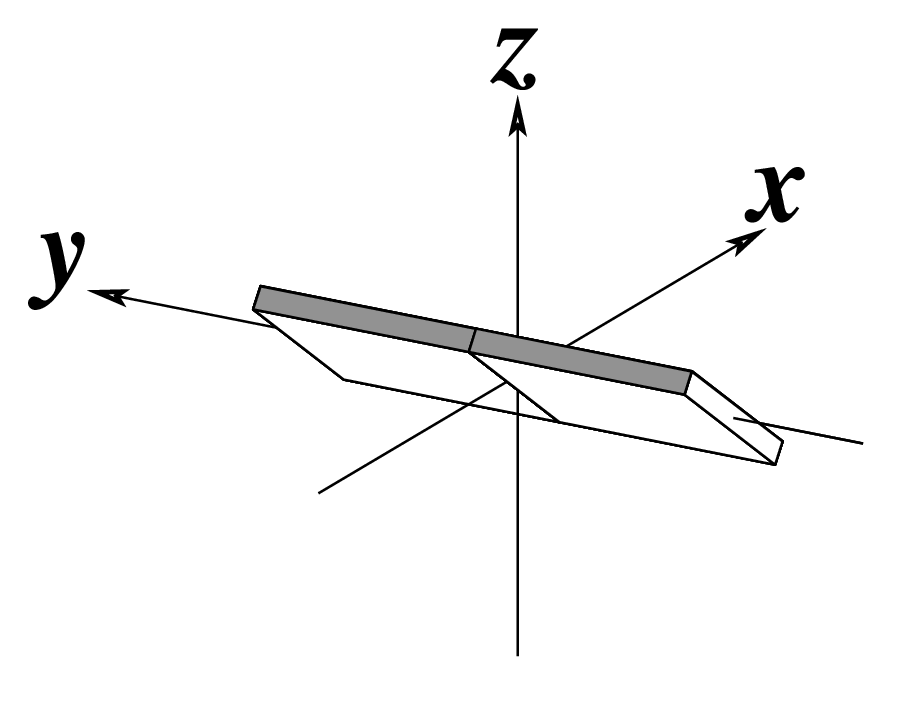
Ever since the 2003 publication of the initial 3DNA Nucleic Acids Research paper (NAR03), the schematic diagrams of base-pair parameters (see figure below) has become quite popular. Over the years, we have received numerous requests for permission to use the figure, or a portion thereof; as an example, the figure has been adopted into a structural biology textbook. In the 2008 3DNA Nature Protocols paper (NP08), we devoted the very first protocol to “create a schematic image for propeller of 45°”.
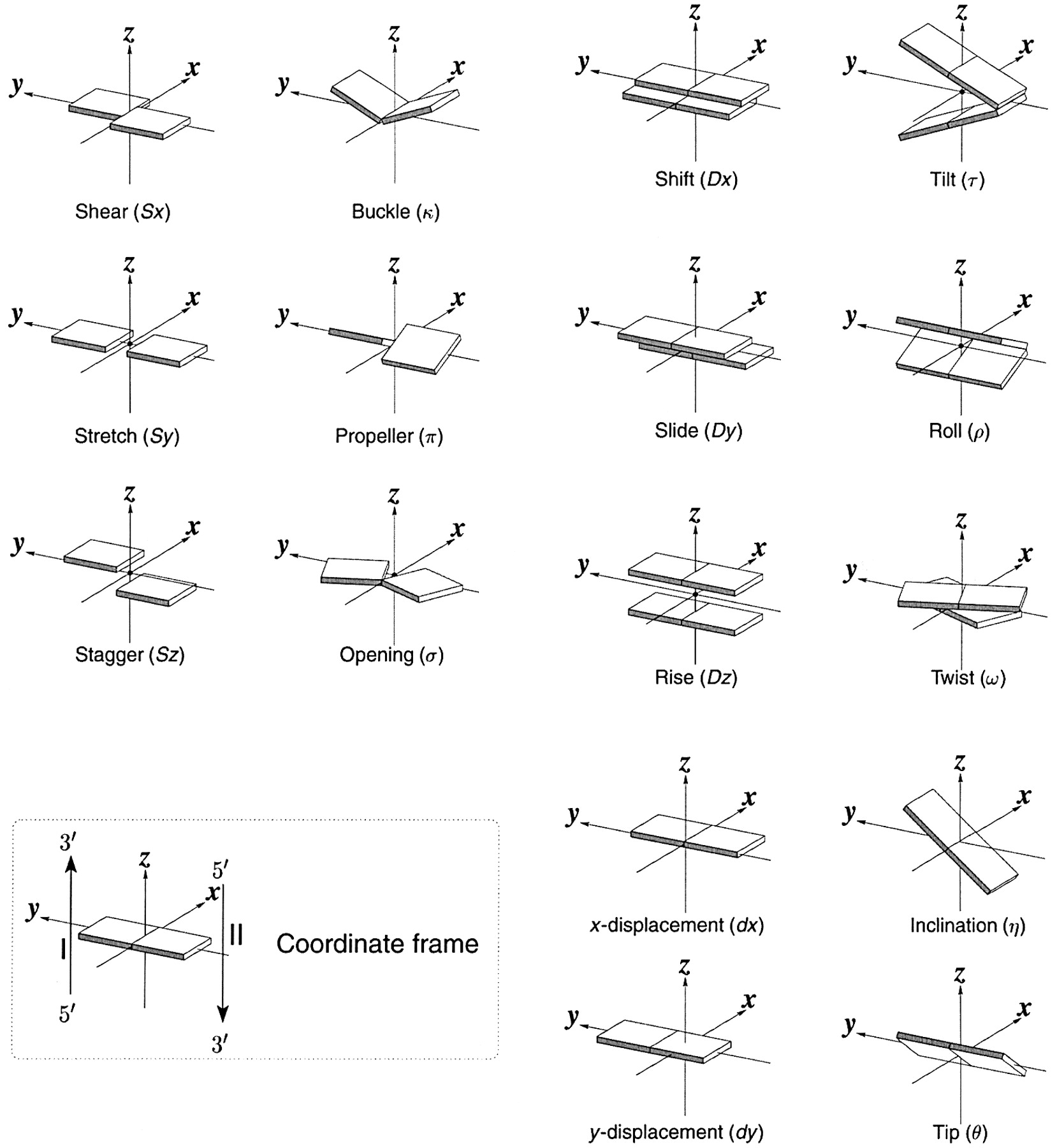
Figure legend taken from Figure 1 of NAR03: Pictorial definitions of rigid body parameters used to describe the geometry of complementary (or non‐complementary) base pairs and sequential base pair steps (19). The base pair reference frame (lower left) is constructed such that the x‐axis points away from the (shaded) minor groove edge of a base or base pair and the y‐axis points toward the sequence strand (I). The relative position and orientation of successive base pair planes are described with respect to both a dimer reference frame (upper right) and a local helical frame (lower right). Images illustrate positive values of the designated parameters. For illustration purposes, helical twist (Ωh) is the same as Twist (ω), formerly denoted by Ω (19,20) and helical rise (h) is the same as Rise (Dz).
I recall spending around two weeks to produce the above figure. Content-wise, the figure was constructed in only a short while; it was the little details that took me most of the time.
Over time, I’ve witnessed numerous versions of such schematic images in publications related to DNA/RNA structures. While looking similar, the schematics differ subtly in the magnitude, orientation and relative scale of illustrated parameters. To the best of my knowledge, only 3DNA provides a pragmatic approach to generate the base-pair schematic diagrams consistently.
To make the schematics more readily accessible, I’ve reproduced a high resolution image (in png format) for each of the 14 parameters shown above. You are welcome to pick and match the diagrams as necessary. If you use any of them in your publications, please cite the 3DNA NAR03 and/or NP08 paper(s).
Note that in the schematic diagrams below, the shaded edge (facing the viewer) denotes the minor-groove side of a base or base pair.
| Shear (Sx) |
Stretch (Sy) |
Stagger (Sz) |
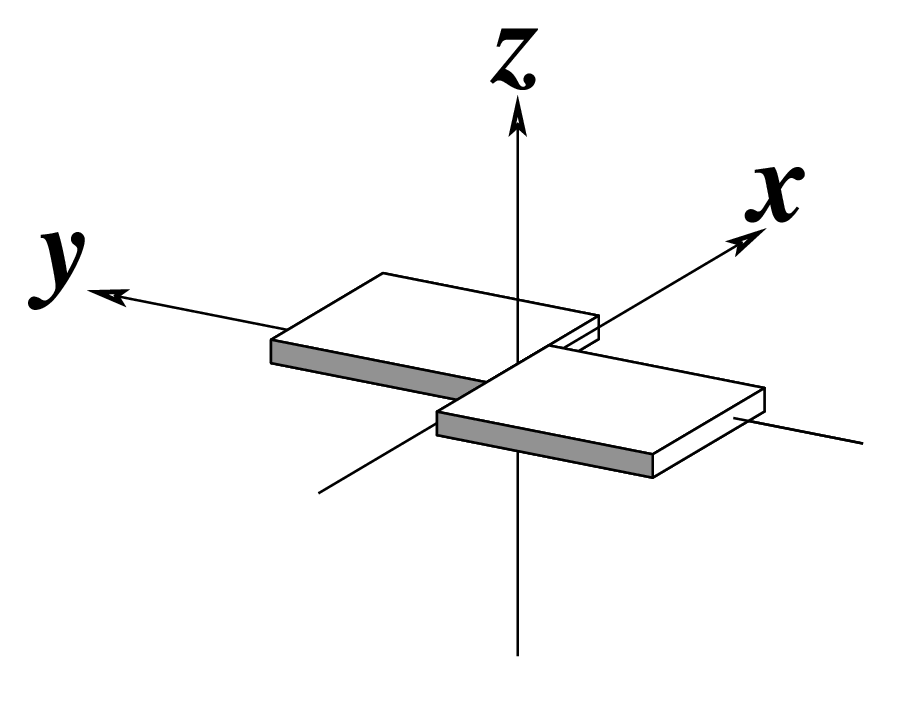 |
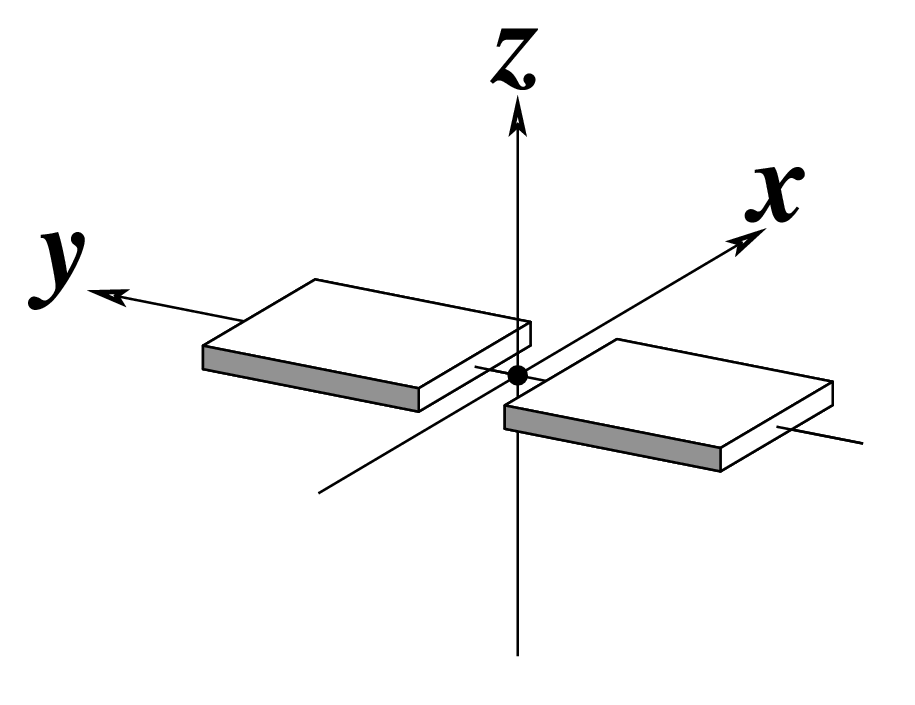 |
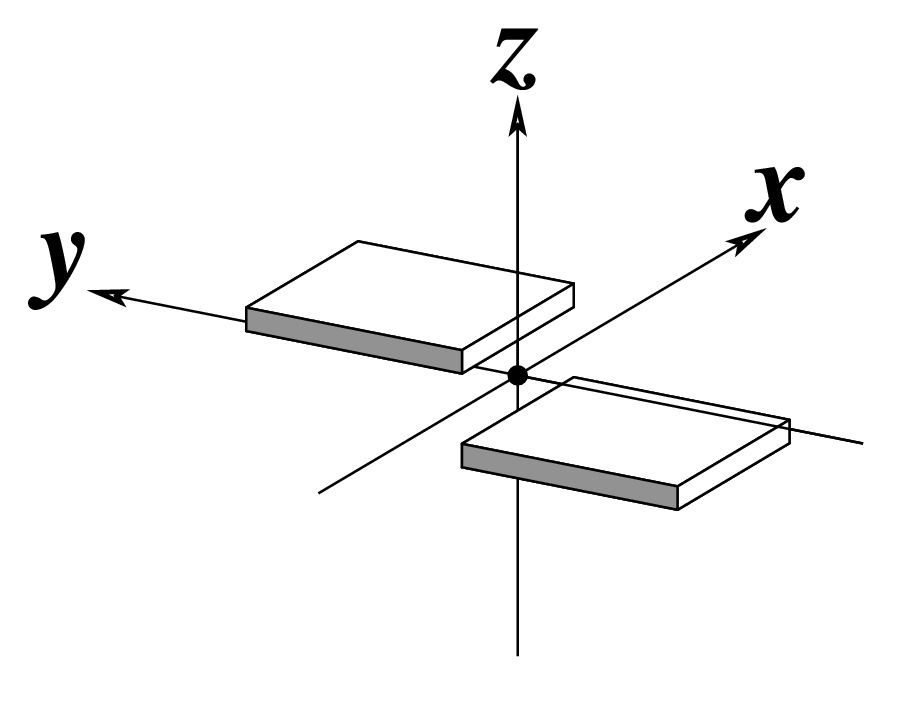 |
| Buckle (κ) |
Propeller (π) |
Opening (σ) |
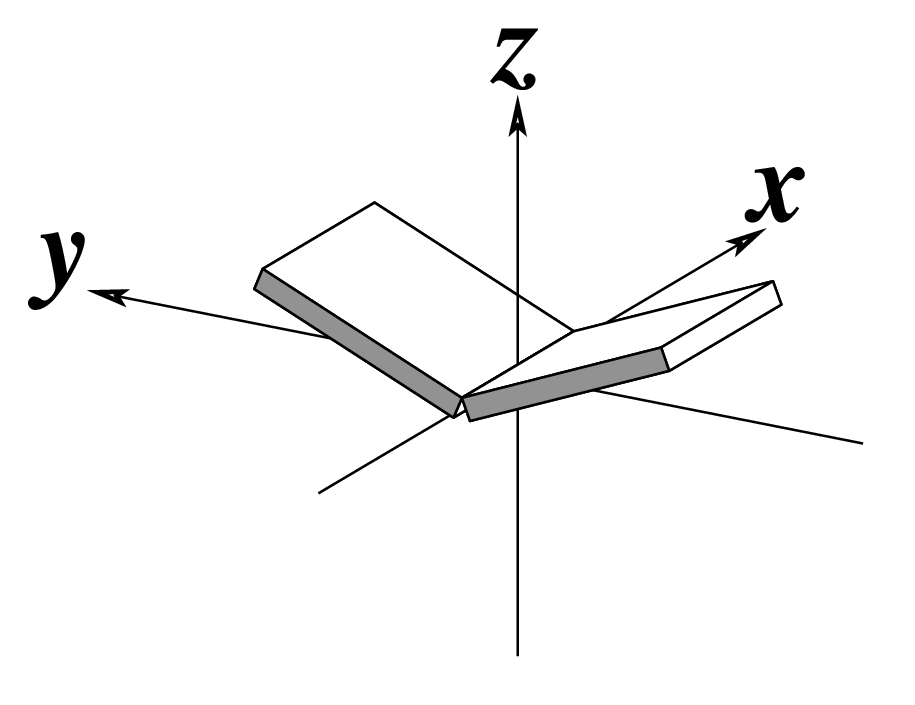 |
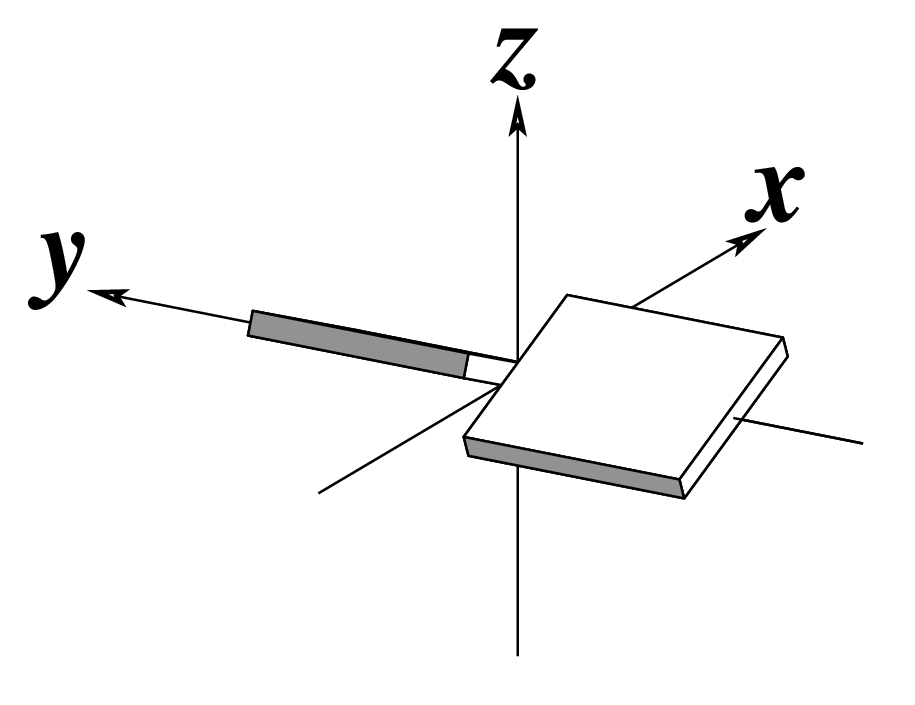 |
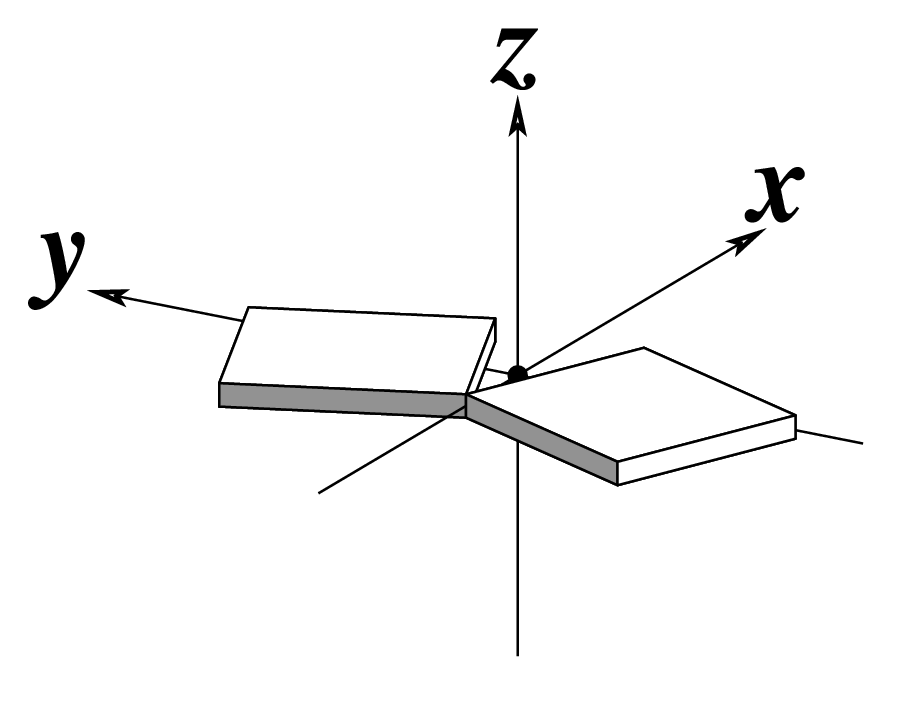 |
| Shift (Dx) |
Slide (Dy) |
Rise (Dz) |
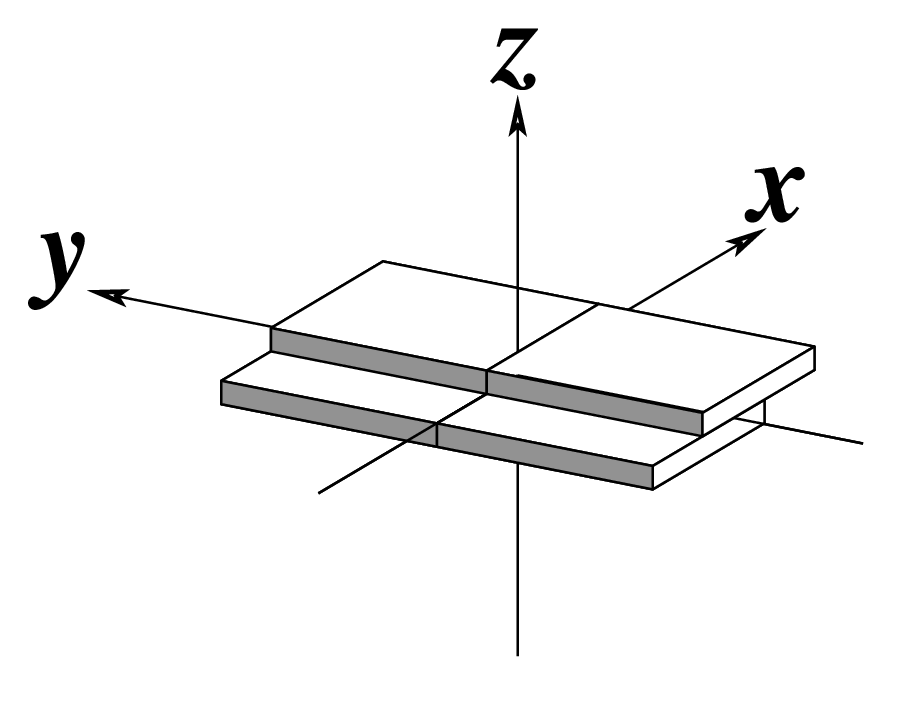 |
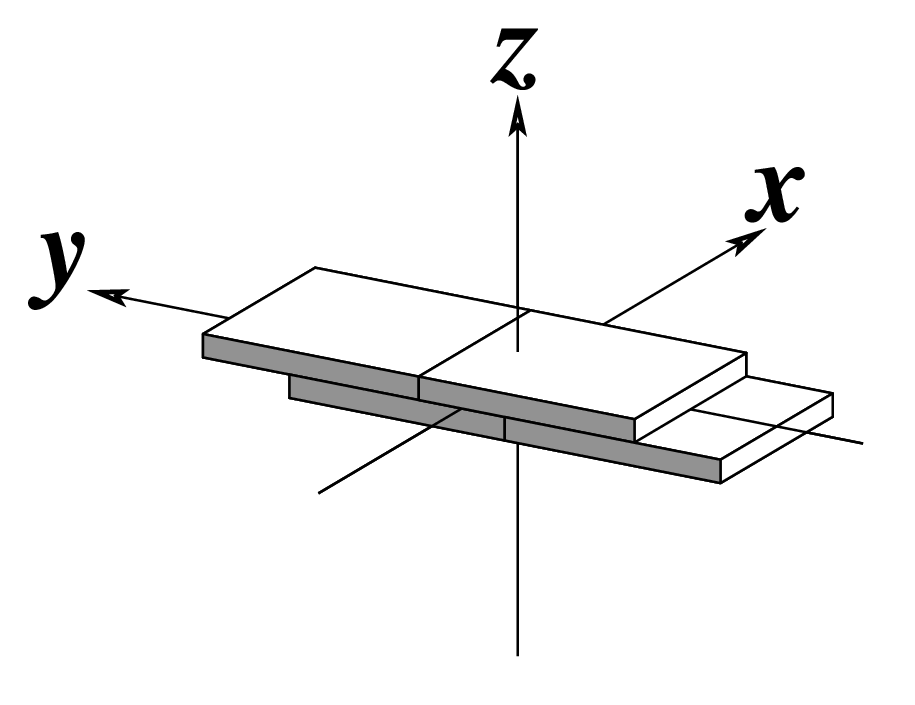 |
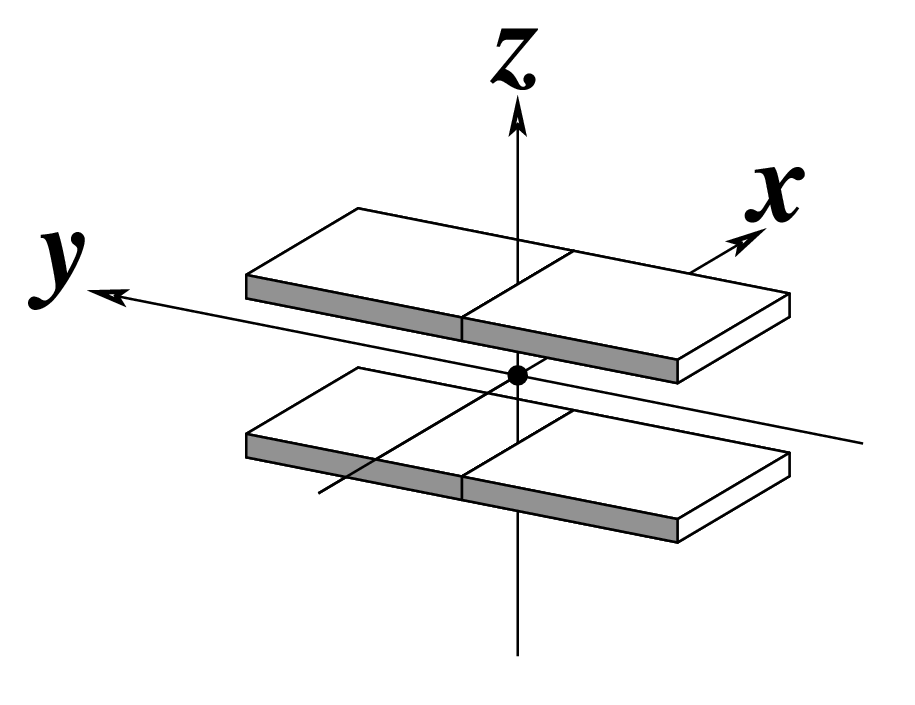 |
| Tilt (τ) |
Roll (ρ) |
Twist (ω) |
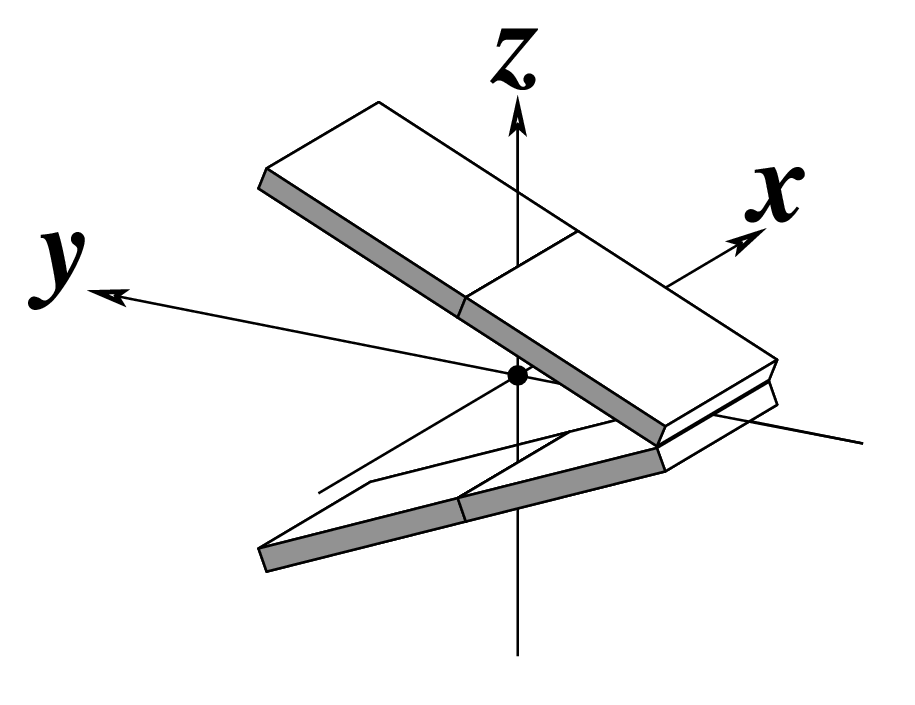 |
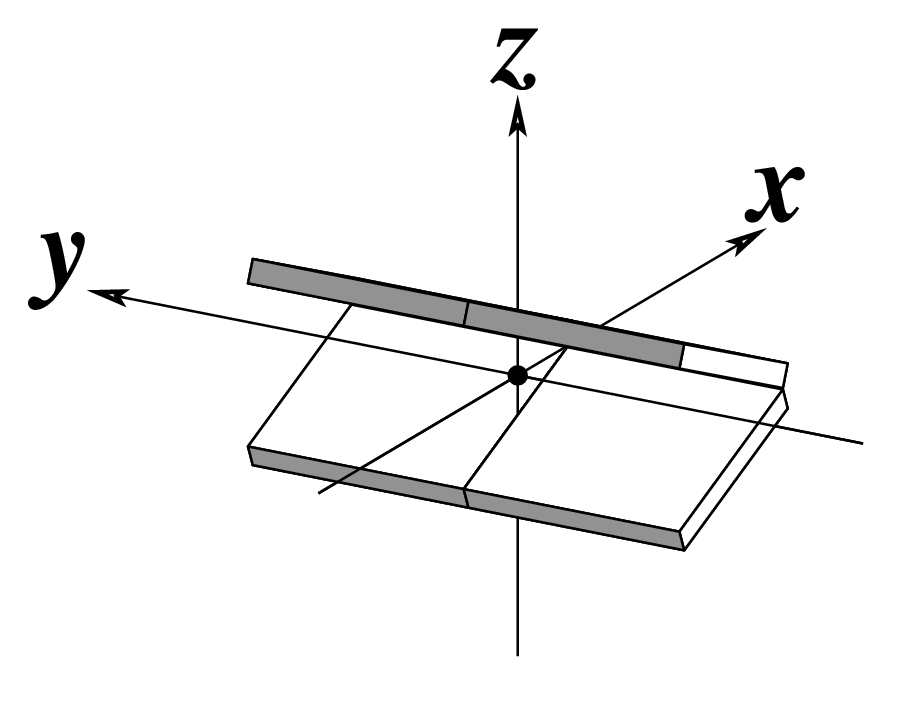 |
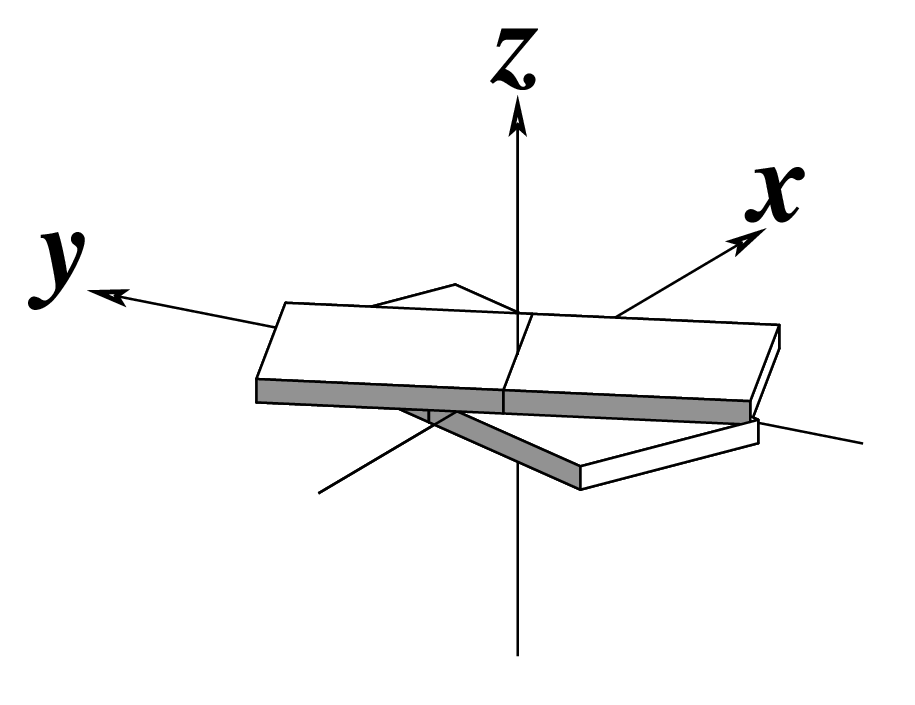 |
| x-displacement (dx) |
y-displacement (dy) |
Helical Rise (h) |
 |
 |
As for Rise above
(for illustration purpose) |
| Inclination (η) |
Tip (θ) |
Helical Twist (Ωh) |
 |
 |
As for Twist above
(for illustration purpose) |













































