Cover images provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
See the 2020 paper titled "DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL" in Nucleic Acids Research and the corresponding Supplemental PDF for details. Many thanks to Drs. Wilma Olson and Cathy Lawson for their help in the preparation of the illustrations.
Details on how to reproduce the cover images are available on the 3DNA Forum.

Structure of the human minor spliceosome pre-B complex (PDB id: 8Y7E; Bai R, Yuan M, Zhang P, Luo T, Shi Y, Wan R. 2024. Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome. Science 383: 1245–1252). The protein–RNA assembly reveals the mechanisms of recognition and recruitment of several small nuclear ribonucleoproteins (snRNPs) involved in the splicing of U12-type introns. The pre-mRNA is depicted by a red ribbon, and the U12 small nuclear RNA (snRNA) by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are shown as gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Human tRNA splicing endonuclease (TSEN) complex bound to pre-tRNAArg (PDB id: 7UXA; Hayne CK, Butay KJ, Stewart ZD, Krahn JM, Perera L, Williams JG, Petrovitch RM, Deterding LJ, Matera AG, Borgnia MJ, Stanley RE. 2023. Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex. Nat Struct Mol Biol 30: 824–833). Cryo-EM structure of the TSEN protein assembly with pre-tRNAArg provides insights into the recognition and splicing of an intron that must be removed from the pre-tRNA before translation. The pre-tRNAArg is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the TSEN subunits are shown as gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Systemic RNA interference defective protein 1 (SID1) in complex with dsRNA (PDB id: 8XC1; Wang R, Cong Y, Qian D, Yan C, Gong D. 2024. Structural basis for double-stranded RNA recognition by SID1. Nucleic Acids Res 52: 6718–6727). The cryo-EM structure provides a major step towards understanding the mechanism of dsRNA recognition by SID1, involving extensive interactions between basic amino-acid residues and the sugar-phosphate backbone. The dsRNA chains are depicted by red, green, blue, and yellow ribbons, with bases and Watson-Crick base pairs represented as color-coded blocks and minor-groove edges colored white: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; SID1 is shown by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Complex of arginyl-tRNA-protein transferase 1 (ATE1) with tRNAArg and a short peptide substrate (PDB id: 8UAU; Lan X, Huang W, Kim SB, Fu D, Abeywansha T, Lou J, Balamurugan U, Kwon YT, Ji CH, Taylor DJ, Zhang Y. 2024. Oligomerization and a distinct tRNA-binding loop are important regulators of human arginyl-transferase function. Nat Commun 15: 6350). The ATE1 homodimer dissociates upon binding the peptide and forms a loop that wraps around tRNAArg. The tRNAArg is depicted by a red ribbon, with bases and Watson–Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; ATE1 is shown by a gold ribbon and the peptide by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of endoribonuclease P (RNase P) in complex with pre-tRNAHis-Ser (PDB id: 8CBK; Meynier V, Hardwick SW, Catala M, Roske JJ, Oerum S, Chirgadze DY, Barraud P, Yue WW, Luisi BF, Tisné C. 2024. Structural basis for human mitochondrial tRNA maturation. Nat Commun 15: 4683). The structure reveals the first step of human mitochondrial tRNA maturation by RNase P, processing the 5′-leader of pre-tRNA. The RNA is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the protein assembly is shown by the gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of a group II intron ribonucleoprotein in the pre-ligation state (PDB id: 8T2R; Xu L, Liu T, Chung K, Pyle AM. 2023. Structural insights into intron catalysis and dynamics during splicing. Nature 624: 682–688). The pre-ligation complex of the Agathobacter rectalis group II intron reverse transcriptase/maturase with intron and 5′-exon RNAs makes it possible to construct a picture of the splicing active site. The intron is depicted by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the 5′-exon is shown by white spheres and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Complex of terminal uridylyltransferase 7 (TUT7) with pre-miRNA and Lin28A (PDB id: 8OPT; Yi G, Ye M, Carrique L, El-Sagheer A, Brown T, Norbury CJ, Zhang P, Gilbert RJ. 2024. Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs. Nat Struct Mol Biol 31: 1426–1438). The RNA-binding pluripotency factor LIN28A invades and melts the RNA and affects the mechanism of action of the TUT7 enzyme. The RNA backbone is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; TUT7 is represented by a gold ribbon and LIN28A by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Cryo-EM structure of the pre-B complex (PDB id: 8QP8; Zhang Z, Kumar V, Dybkov O, Will CL, Zhong J, Ludwig SE, Urlaub H, Kastner B, Stark H, Lührmann R. 2024. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 630: 1012–1019). The pre-B complex is thought to be critical in the regulation of splicing reactions. Its structure suggests how the cross-exon and cross-intron spliceosome assembly pathways converge. The U4, U5, and U6 snRNA backbones are depicted respectively by blue, green, and red ribbons, with bases and Watson-Crick base pairs shown as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are represented by gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the Hendra henipavirus (HeV) nucleoprotein (N) protein-RNA double-ring assembly (PDB id: 8C4H; Passchier TC, White JB, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN. 2024. The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Sci Rep 14: 14099). The HeV N protein adopts a bi-lobed fold, where the N- and C-terminal globular domains are bisected by an RNA binding cleft. Neighboring N proteins assemble laterally and completely encapsidate the viral genomic and antigenomic RNAs. The two RNAs are depicted by green and red ribbons. The U bases of the poly(U) model are shown as cyan blocks. Proteins are represented as semitransparent gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the helicase and C-terminal domains of Dicer-related helicase-1 (DRH-1) bound to dsRNA (PDB id: 8T5S; Consalvo CD, Aderounmu AM, Donelick HM, Aruscavage PJ, Eckert DM, Shen PS, Bass BL. 2024. Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA. eLife 13: RP93979. Cryo-EM structures of Dicer-1 in complex with DRH-1, RNAi deficient-4 (RDE-4), and dsRNA provide mechanistic insights into how these three proteins cooperate in antiviral defense. The dsRNA backbone is depicted by green and red ribbons. The U-A pairs of the poly(A)·poly(U) model are shown as long rectangular cyan blocks, with minor-groove edges colored white. The ADP ligand is represented by a red block and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Moreover, the following 30 [12(2021) + 12(2022) + 6(2023)] cover images of the RNA Journal were generated by the NAKB (nakb.org).
Cover image provided by the Nucleic Acid Database (NDB)/Nucleic Acid Knowledgebase (NAKB; nakb.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Standard nitrogenous bases in DNA and RNA (A, C, G, T, and U) are aromatic compounds, each with a planar geometry. In the analyses of three-dimensional (3D) nucleic acid structures, the planar bases are normally taken as rigid bodies. The relative geometry of the two bases in base pair (bp) can then be rigorously quantified by six rigid-body parameters (see figure below). The three translations along the _x_-, _y_- and _z_-axes are termed Shear, Stretch, and Stagger, respectively. The three corresponding rotations are called Buckle, Propeller (twist), and Opening.

3DNA is unique with its coupled analyze and rebuild programs. The former calculates six bp parameters given 3D atomic coordinates (in PDB or PDBx/mmCIF format), while the later takes a set of such parameters to generate the corresponding structure. The rigor of the description can be easily verified in two equivalent ways: the close to zero root-mean-square deviation (RMSD) between the rebuilt structure and the original coordinates, after a least-squares superposition; or the identical six bp parameters when the rebuilt structure is analyzed.
As is often the case, a concrete example would make the point clear. Here I am using the reverse Hoogsteen (rHoogsteen) bp between U8 and A14 (see image below) in the yeast phenylalanine tRNA (1ehz) as an example. The PDB atomic coordinates of the U8–A14 rHoogsteen pair, excluding backbone atoms except for C1′, is stored in file 1ehz-U8-A14.pdb.

find_pair 1ehz-U8-A14.pdb stdout | analyze stdin
# bp parameters in file '1ehz-U8-A14.out'
# also generated 'bp_step.par' for rebuilding below
rebuild -atomic bp_step.par 1ehz-U8-A14-3DNA.pdb
# rmsd is 0.044 Å between '1ehz-U8-A14.pdb' and '1ehz-U8-A14-3DNA.pdb'
find_pair 1ehz-U8-A14-3DNA.pdb stdout | analyze stdin
# bp parameters of the rebuilt structure in '1ehz-U8-A14-3DNA.out'
rebuild -atomic bp_step.par 1ehz-U8-A14-3DNA-new.pdb
# rmsd is 0 Å between '1ehz-U8-A14-3DNA.pdb' and '1ehz-U8-A14-3DNA-new.pdb'
Note that the above commands should be performed in order, since the file bp_step.par is overwritten after each analyze run. For your verification, here are the links to the five files:
The 0.044 Å rmsd between the original PDB coordinates in 1ehz-U8-A14.pdb and the 3DNA rebuilt structure in 1ehz-U8-A14-3DNA.pdb is due to the slight non-planarity of experimental bases. The rmsd is 0 between the two rounds of 3DNA rebuilt structures, 1ehz-U8-A14-3DNA.pdb and 1ehz-U8-A14-3DNA-new.pdb, as expected.
The bp parameters in 1ehz-U8-A14.out and 1ehz-U8-A14-3DNA.out are identical, as expected, and they are shown below.
Local base-pair parameters
bp Shear Stretch Stagger Buckle Propeller Opening
1 U-A 4.14 -1.91 0.77 -4.62 12.12 -103.09
Running DSSR on 1ehz-U8-A14.pdb gives the following results. Note that the six bp parameters (last row prefixed with bp-pars) are the exactly same as in 3DNA — we are consistent.
# x3dna-dssr -i=1ehz-U8-A14.pdb --more
List of 1 base pair
nt1 nt2 bp name Saenger LW DSSR
1 A.U8 A.A14 U-A rHoogsteen 24-XXIV tWH tW-M
[n/a(n/a) ---- lambda=28.3] [n/a(n/a) ---- lambda=21.5]
d(C1'-C1')=9.63 d(N1-N9)=7.06 d(C6-C8)=6.00 tor(C1'-N1-N9-C1')=174.4
H-bonds[2]: "O2(carbonyl)-N6(amino)[3.00],N3(imino)-N7[2.74]"
interBase-angle=12.97 Simple-bpParams: Shear=4.28 Stretch=1.55 Buckle=-11.8 Propeller=5.4
bp-pars: [4.14 -1.91 0.77 -4.62 12.12 -103.09]
As mentioned in the recent DSSR paper:
As in 3DNA (6,7), DSSR takes advantage of the six standard base-pair parameters––three translations (Shear, Stretch, Stagger) and three rotations (Buckle, Propeller, Opening)––to quantify the relative spatial position and orientation of any two interacting bases rigorously. Among the six parameters, only Shear, Stretch, and Opening are critical for characterizing different types of pairs. Buckle, Propeller and Stagger, on the other hand, describe the nonplanarity of a given pair (6). By virtue of the definition of the standard base reference frame, Shear, Stretch, and Opening are all close to zero for Watson-Crick pairs. Moreover, every other type of pair has a set of characteristic parameters. For example, the wobble G–U pair is characterized by an average Shear of –2.2 Å, and the Hoogsteen A+U pair is distinguished by a Stretch of approximately –3.5 Å and an Opening of near 66º.
In a follow-up post, I will talk about the “simple” bp parameters (Simple-bpParams in the above DSSR output list) recently introduced into DSSR — stay tuned!

As of DSSR v1.3.0-2015aug27, the --json option is available for producing analysis results that is strictly compliant with the JSON data exchange format. The JSON file contains numerous DSSR-derived structural features, including those in the default main output, backbone torsions in dssr-torsions.txt, and a detailed list of hydrogen bonds.
According to the official JSON website:
JSON (JavaScript Object Notation) is a lightweight data-interchange format. It is easy for humans to read and write. It is easy for machines to parse and generate. It is based on a subset of the JavaScript Programming Language… JSON is a text format that is completely language independent… These properties make JSON an ideal data-interchange language.
Indeed, the JSON output file makes DSSR readily accessible for integration with other bioinformatics tools or normal usages from the command line. Using the classic yeast phenylalanine tRNA 1ehz as an example (1ehz.pdb), let’s go over some simple use-cases. Note that the following examples take advantage of jq, a lightweight and flexible command-line JSON processor.
x3dna-dssr -i=1ehz.pdb --json -o=1ehz-dssr.json
jq . 1ehz-dssr.json # reformatted for pretty output
x3dna-dssr -i=1ehz.pdb --json | jq . # the above 2 steps combined
With 1ehz-dssr.json in hand, we can easily extract DSSR-derived structural features of interest:
jq .pairs 1ehz-dssr.json # list of 34 pairs
jq .multiplets 1ehz-dssr.json # list of 4 base triplets
jq .hbonds 1ehz-dssr.json # list of hydrogen bonds
jq .helices 1ehz-dssr.json
jq .stems 1ehz-dssr.json
# list of nucleotide parameters, including torsion angles and suites
jq .ntParams 1ehz-dssr.json
# list of 14 modified nucleotides
jq '.ntParams[] | select(.is_modified)' 1ehz-dssr.json
# select nucleotide id, delta torsion, sugar puckering and cluster of suite name
jq '.ntParams[] | {nt_id, delta, puckering, cluster}' 1ehz-dssr.json
# same selection as above, but in 'Comma Separated Values' format
jq -r '.ntParams[] | [.nt_id, .delta, .puckering, .cluster] | @csv' 1ehz-dssr.json
Here is the result of running jq (v1.5) to select multiplets:
# jq .multiplets 1ehz-dssr.json
[
{
"index": 1,
"num_nts": 3,
"nts_short": "UAA",
"nts_long": "A.U8,A.A14,A.A21"
},
{
"index": 2,
"num_nts": 3,
"nts_short": "AUA",
"nts_long": "A.A9,A.U12,A.A23"
},
{
"index": 3,
"num_nts": 3,
"nts_short": "gCG",
"nts_long": "A.2MG10,A.C25,A.G45"
},
{
"index": 4,
"num_nts": 3,
"nts_short": "CGg",
"nts_long": "A.C13,A.G22,A.7MG46"
}
]
With the JSON file, DSSR can now be connected with the bioinformatics community in a ‘structured’ way, with a clearly delineated boundary. Now I can enjoy the freedom of refining the default main output format, without worrying too much about breaking third-party parsers. Moreover, I no longer need to write an adapter for each integration of DSSR with other tools. So nice!
For your reference, here is the output file 1ehz-dssr.json. It may be possible that the identifiers (names) of the JSON output will be refined in the next few iterations. I welcome your comments to make the DSSR-derived JSON better suite your needs.

It is a great pleasure to note that a paper titled DSSR, an integrated software tool for dissecting the spatial structure of RNA has recently been published in Nucleic Acids Research (NAR). Co-authored by Harmen Bussemaker, Wilma Olson and me (a team with a unique combination of complementary expertise), this DSSR paper represents another solid piece of work that I feel proud of. In contrast to our previous GpU dinucleotide platform paper focusing on results, and the two major 3DNA papers concentrating on methods, the current NAR article describes significant scientific findings that are enabled by the novel analysis algorithms implemented in the program. Moreover, DSSR introduces an appealing and highly informative “cartoon-block” representation of RNA structures that combines PyMOL cartoon schematics with 3DNA base color-coded rectangular blocks.
The abstract of the paper is quoted below:
Insight into the three-dimensional architecture of RNA is essential for understanding its cellular functions. However, even the classic transfer RNA structure contains features that are overlooked by existing bioinformatics tools. Here we present DSSR (Dissecting the Spatial Structure of RNA), an integrated and automated tool for analyzing and annotating RNA tertiary structures. The software identifies canonical and noncanonical base pairs, including those with modified nucleotides, in any tautomeric or protonation state. DSSR detects higher-order coplanar base associations, termed multiplets. It finds arrays of stacked pairs, classifies them by base-pair identity and backbone connectivity, and distinguishes a stem of covalently connected canonical pairs from a helix of stacked pairs of arbitrary type/linkage. DSSR identifies coaxial stacking of multiple stems within a single helix and lists isolated canonical pairs that lie outside of a stem. The program characterizes ‘closed’ loops of various types (hairpin, bulge, internal, and junction loops) and pseudoknots of arbitrary complexity. Notably, DSSR employs isolated pairs and the ends of stems, whether pseudoknotted or not, to define junction loops. This new, inclusive definition provides a novel perspective on the spatial organization of RNA. Tests on all nucleic acid structures in the Protein Data Bank confirm the efficiency and robustness of the software, and applications to representative RNA molecules illustrate its unique features. DSSR and related materials are freely available at http://home.x3dna.org/.
During the review process, we are delighted that the referees confirmed the claim that we made in the cover letter: “We would also like to emphasize that our reported results are easily verifiable, and we assure rigorous reproducibility of the data and figures described in this article.” Now that the paper has been published, as a follow-up, I’ve made available all the scripts and data files associated with the paper in a new section DSSR-NAR paper on the 3DNA Forum. The DSSR User Manual has also been updated with additional, previously undocumented, auxiliary options.
Overall, it took me more than ten days to create the 19 posts in the DSSR-NAR paper section and to revise the DSSR User Manual, along with other minor refinements for consistency. During the process, I’ve tried to make the scripts and data files self-contained for wide accessibility and easy understanding.
Any interested party should now be able to reproduce the table and figures (including the supplementary data) reported in the article. Moreover, with the additional details given in the post RNA cartoon-block representations with PyMOL and DSSR, one can easily generate similar schematic images as shown below:
I feel confident to claim that the results reported in our DSSR paper are reproducible. If you have issues related to the paper, please post them on the 3DNA Forum. I strive to respond promptly to any questions asked there.
In summary, DSSR is an integrated computational tool, designed from the bottom up to streamline the analysis of RNA three-dimensional structures. It is built upon my extensive experience in supporting 3DNA, growing knowledge of RNA structures, and refined programming skills. DSSR has a combined set of functionalities well beyond the scope of any known specialized resources. The program may well serve as a cornerstone for RNA structural bioinformatics and will benefit a broad range of possible applications.

Nowadays, “big data” and “big science” are hot topics. They all sound good and certainly come about for a reason. Yet, to transform data to information to knowledge to understanding to wisdom, sophisticated software tools are required. The programs can be big and complicated, or small and self-contained, fitting different purposes. As long as they can get the claimed job done in a robust fashion, size should not be a concern.
Over the years, however, I have seen a trend of bloated software with many (fragile) dependencies in bioinformatics. Some tools are so picky and hard to use/maintain that instead of serving, they become sort of a master. As a more representative example, I recently tried to install an open-source software associated with a paper published just a few years ago in a leading journal. The software has only a few dependencies, yet some of them have already become obsolete. I spent hours each time, on Mac OS X and two versions of Ubuntu Linux, but failed to get it running properly (always abort with error messages). The download page hosting the software has been inactive since around the publication of the paper. Presumably, the PhD student or postdoc who wrote the code had left the lab, and with a paper published, all is done!
As an active practitioner of bioinformatics for well over a decade, I can confidently claim to be well above average in familiarity with Linux/Mac OS X and associated shell programming and make etc tools, and various common scripting and compiled programming languages. Yet, once in a while, I get frustrated when I try to download and install a software tool attached to a paper I am interested in. As I see it, the vast majority of software programs from research labs are publication-oriented — as long a paper is published, it is finished.
From my experience, I always see software as engineering. It needs careful design and great attention to meticulous details. A sophisticated piece of scientific software is a combination of science and engineering. Expertise in domain knowledge is a must, and refined skills in computer programming is indispensable. The DSSR program I created and continuously refined over the past three years represents what a scientific software should be in my believe.
Among other unique features, DSSR is tiny (< 1mb), self-contained (without run-time dependencies) and runs on Windows, Mac OS X, and Linux. Getting DSSR up and running should take only minutes by any one with basic familiarity of common computer systems. I have no doubt that the beauty of being small as represented by DSSR will be gradually appreciated by the community.

Over the past few weeks, I’ve had the pleasure to talk to Thomas Holder, the PyMOL Principal Developer at Schrödinger, on possible integration of DSSR into PyMOL. On Tuesday April 21, 2015, I wrote to Thomas:
Last year, I had the please to collaborate with Dr. Robert Hanson to integrate DSSR into Jmol, see
http://chemapps.stolaf.edu/jmol/jsmol/dssr.htm. I am wondering if you have any interest in connecting DSSR to PyMOL. This will not only benefit both parties, but also bring elaborate analyses of RNA structures to the general audience. As you may be aware, RNA is becoming increasing important, yet the field of RNA structural bioinformatics is lagging (far) behind that of proteins.
After a few meet-ups, we all agree that the DSSR-PyMOL integration project would be meaningful/significant for RNA structural bioinformatics. Moreover, the community not only can benefit from the end result, but also should be able to make direct contributions through the process. On Friday May 08, 2015, Thomas sent out the following open invitation, titled Someone interested in writing a DSSR plugin for PyMOL?, to the PyMOL mailing list:
Is anyone interested in writing a DSSR plugin for PyMOL? DSSR is an integrated software tool for Dissecting the Spatial Structure of RNA (http://x3dna.bio.columbia.edu/docs/dssr-manual.pdf). Among other things, DSSR defines the secondary structure of RNA from 3D atomic coordinates in a way similar to DSSP does for proteins. Most of its output could be translated 1:1 into PyMOL selections, making it available for coloring and other selection based features. A PyMOL plugin could act as a wrapper which runs DSSR for an object or atom selection. Xiang-Jun Lu, the author of DSSR, is also working on base pair visualization (see http://home.x3dna.org/articles/seeing-is-understanding-as-well-as-believing), similar to (but more advanced) what’s already available from 3DNA (http://pymolwiki.org/index.php/3DNA).
Xiang-Jun would be happy to collaborate with someone who has experience with Python and the PyMOL API for writing an extension or plugin. Please contact me if this sounds appealing to you.
Get DSSR from http://home.x3dna.org/
See it hooked up with JSmol: http://chemapps.stolaf.edu/jmol/jsmol/dssr.htm
If you are self-motivated, care about software quality, have expertise in writing PyMOL plugin, and feel the pain in RNA structural analysis/visualization with currently available tools, now it is the time to make a difference. The DSSR/PyMOL project would ideally be composed of a team of dedicated practitioners with complementary skills. We will communicate mostly via email or online forum, in a presumably open and highly interactive way. By working on the project, you will be able to sharpen your skills and make new friends. The end product would not only make RNA structural bioinformatics easier for yourself but also benefit the community at large.

The v1.2.1 (2015feb01) release of DSSR contains a new functionality to characterize the so-called H-type pseudoknots. In this classical and most common type of pseudoknots, nucleotides from a hairpin loop form Watson-Crick base pairs with a single-stranded region outside of the hairpin to create another (adjacent) stem, as shown in the following illustration (taken from the Huang et al. paper A heuristic approach for detecting RNA H-type pseudoknots).

Normally, L2 is absent (i.e., with zero nucleotides) due to direct coaxial stacking of the two stems. An example output of DSSR on 1ymo (a human telomerase RNA pseudoknot) is shown below:
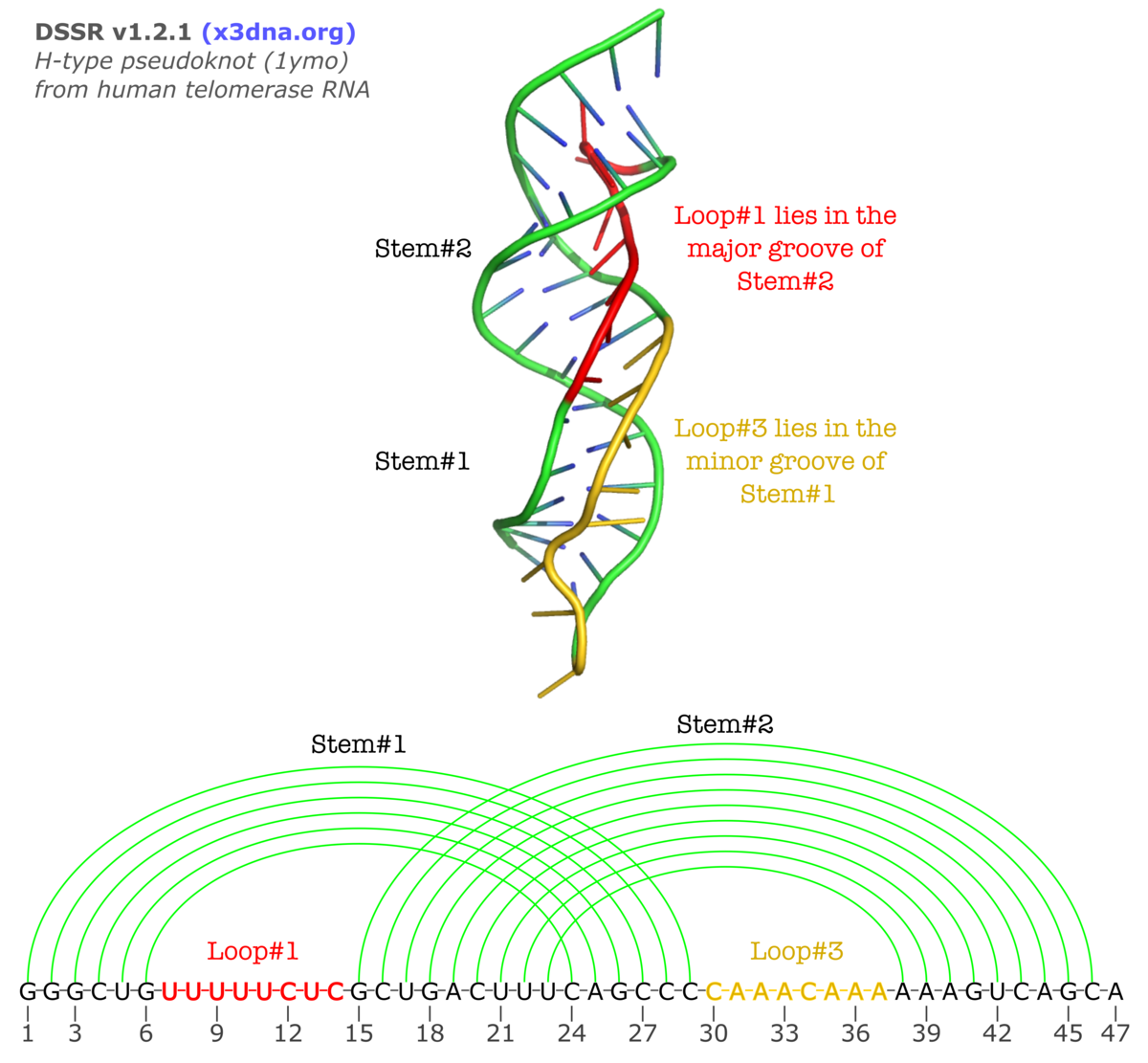
The corresponding sections from DSSR output are:
****************************************************************************
List of 3 H-type pseudoknot loop segments
1 stem#1(hairpin#1) vs stem#2(hairpin#2) L1 groove=MAJOR nts=8 UUUUUCUC U7,U8,U9,U10,U11,C12,U13,C14
2 stem#1(hairpin#1) vs stem#2(hairpin#2) L2 groove=----- nts=0
3 stem#1(hairpin#1) vs stem#2(hairpin#2) L3 groove=minor nts=8 CAAACAAA C30,A31,A32,A33,C34,A35,A36,A37
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>1ymo-1-A #1 nts=47 [chain] RNA
GGGCUGUUUUUCUCGCUGACUUUCAGCCCCAAACAAAAAAGUCAGCA
[[[[[[........(((((((((]]]]]]........))))))))).
Checking against the three-dimensional image and the secondary structure in linear form shown above, the meaning of the new section should be obvious. If you want to see more details, click the link to the DSSR-output file on 1ymo.

Recently I came across the following two citations to DSSR:
Base pair types were annotated with RNAview (45,46). Hydrogen bonds were annotated manually and with the help of DSSR of the 3DNA package (47,48). Helix parameters were obtained using the Curves+ web server (49). Structural figures were prepared using PyMol (50).
It is interesting to note that DSSR is cited here for its identification of hydrogen bonds, not its annotation of base pairs, among many other features. The simple geometry-based H-bonding identification algorithm, originally implemented in find_pair/analyze of 3DNA (and adopted by RNAView) and highly refined in DSSR, works well for nucleic acid structures. With the --get-hbonds option, users can now use DSSR as a tool just for its list of H-bonds outside of the program.
All figures were generated using PyMOL (60) or Chimera (48). The secondary structure diagram of the human mitoribosomal RNA was prepared by extracting base pairs from the model using DSSR (61). The secondary structure diagram was drawn in VARNA (62) and finalized in Inkscape.
I am very pleased to see that DSSR was cited for its ‘intended’ use in this important piece of work from a leading laboratory in structural biology. In the middle of last November (2013), I was approached by the lead author for proper citation of DSSR, and I suggested the two 3DNA papers. As far as I can remember, this was the first time I received such a question on DSSR citation. It prompted to write a FAQ entry in the DSSR User Manual, titled “How to cite DSSR?”. Hopefully, this citation issue will be gone in the near future.
Over the past two years, I’ve devoted significant efforts to make DSSR a handy tool for RNA structural bioinformatics; it certainly represents my view as to what a scientific software program should be like. As time passes by, DSSR is becoming increasingly sophisticated and citations to DSSR can only be higher.

Recently, PDB begins to release atomic coordinates of large (ribosomal) structures in mmCIF format. For nucleic-acid-containing structures, the largest one so far is 4v4g, the crystal structure of five 70S ribosomes from Escherichia coli in complex with protein Y. It is assembled from ten PDB entries (1voq, 1vor, 1vos, 1vou, 1vov, 1vow, 1vox, 1voy, 1voz, 1vp0), consisting of 22,345 nucleotides, and a total of 717,805 atoms.
This humongous structure poses no problems to DSSR at all, as shown below.
Command: x3dna-dssr -i=4v4g.cif -o=4v4g.out
Processing file '4v4g.cif' [4v4g]
total number of base pairs: 9277
total number of multiplets: 918
total number of helices: 1099
total number of stems: 1221
total number of isolated WC/wobble pairs: 603
total number of atom-base stacking interactions: 1736
total number of hairpin loops: 504
total number of bulges: 170
total number of internal loops: 775
total number of junctions: 214
total number of non-loop single-stranded segments: 429
total number of kissing loops: 5
total number of A-minor (type I and II) motifs: 100
total number of ribose zippers: 58 (1159)
total number of kink turns: 39
Time used: 00:00:10:45
It took less than 11 minutes to run on an iMac (and nearly 14 minutes on a Ubuntu Linux machine). Given the

From early on, 3DNA and DSSR have native support of modified nucleotides. The currently distributed baselist.dat file with 3DNA contains over 700 entries. As of v1.1.4-2014aug09, a new section has been added to DSSR to list explicitly the modified nucleotides in an analyzed structure.
Using the 76-nucleotide long yeast phenylalanine tRNA (1ehz) as an example, the pertinent section in DSSR output is as below.
List of 11 types of 14 modified nucleotides
nt count list
1 1MA-a 1 A.1MA58
2 2MG-g 1 A.2MG10
3 5MC-c 2 A.5MC40,A.5MC49
4 5MU-t 1 A.5MU54
5 7MG-g 1 A.7MG46
6 H2U-u 2 A.H2U16,A.H2U17
7 M2G-g 1 A.M2G26
8 OMC-c 1 A.OMC32
9 OMG-g 1 A.OMG34
10 PSU-P 2 A.PSU39,A.PSU55
11 YYG-g 1 A.YYG37
So 1ehz has 14 modified nucleotides of 11 different type, as listed in the following rows after the header line. The meaning of each column should be obvious. For example, the third row means that 5MC (5-methylcytidine, abbreviated as 'c' in 1-letter code) occurs twice, identified as A.5MC40 and A.5MC49, respectively.
With the 3-letter id, one can search the RCSB ligand database for more information about a specified modified nucleotide. The URL would be like this, using pseudouridine (PSU) as an example, https://www.rcsb.org/ligand/PSU.
It is hoped that the newly added section, put at the very top of DSSR output, will draw more attention to modified nucleotides.