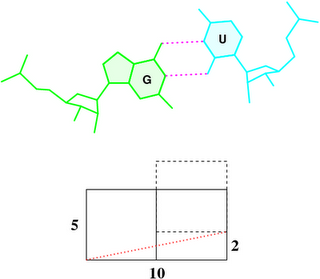
In the standard base reference frame report, a whole section is devoted to the discussion of intrinsic correlations between base-pair and dimer step parameters (see figure below). Among the four sets of associations, the effect of Δbuckle (difference in consecutive base-pair buckles) on rise is most noticeable and easiest to understand. The Δshear vs. twist relationship is similarly significant, due to its close connection to the wobble G–T/G–U pair; yet the concept is less comprehensible, especially to occasional 3DNA users. This post aims to address the issue of how Δshear effects twist.
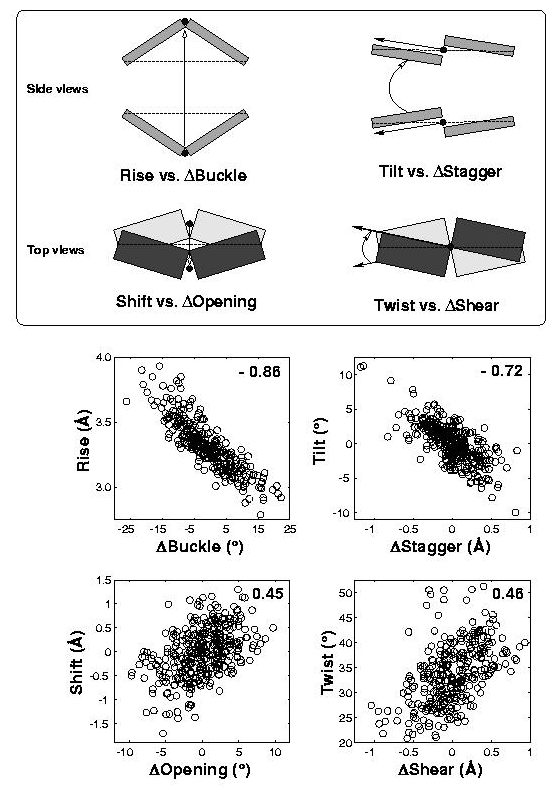
Under the standard base reference frame used in 3DNA, the wobble base-pair has a ~2.0 Å shear: the displacement is positive for U–G, and negative for G–U [see figure below, examples selected from 5S rRNA (chain 9) U82–G100 and G83–U99 of the Haloarcula marismortui large ribosomal subunit, PDB id: 1jj2].
 |
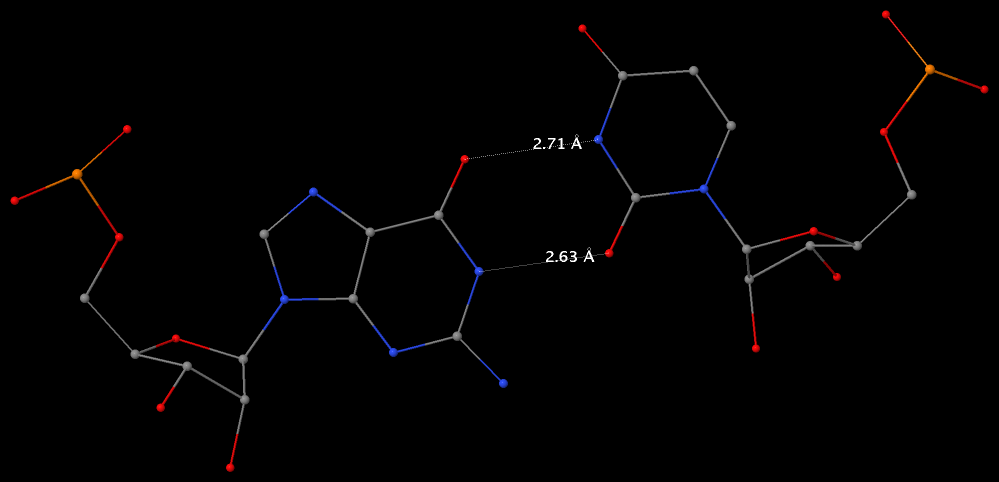 |
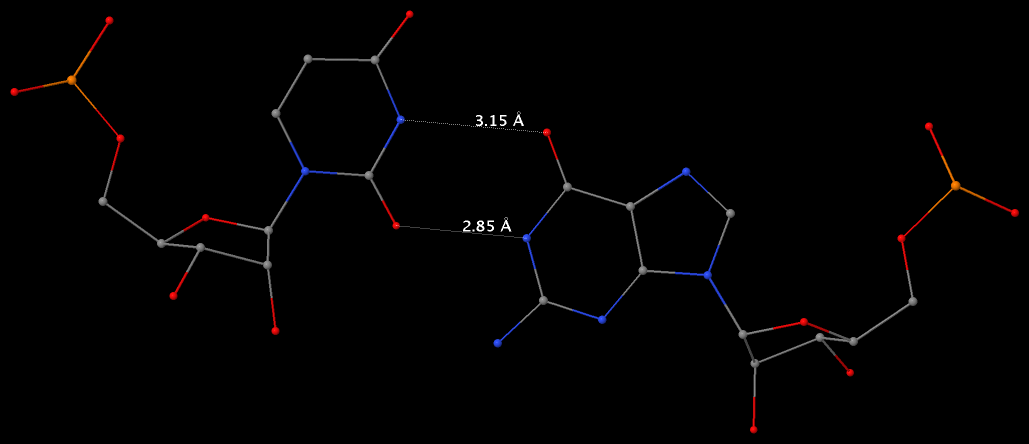
As noted in the section “treatment of non-Watson–Crick base pairing motifs” of the 3DNA Nucleic Acids Research paper (2003), “Large Shear of the G–U wobble base pair influences the calculated but not the ‘observed’ Twist. The 3DNA numerical values of Twist [of the C7G8·U12G13 and G8C9·G11U12 dimer steps of the Escherichia coli tRNAAsp x-ray crystal structure (PDB id: 485d)], 20° (top) and 43° (bottom), differ from the visualization of nearly equivalent Twist suggested by the angle between successive C1′···C1′ vectors (finely dotted lines).”
To make it clear why that’s the case, the figure below shows a G–U wobble pair in atomic representation (top), and a schematic base pair rectangular block of dimension 10×5 (Å, bottom). A shear of –2 Å moves U upwards, as outlined by the dashed rectangle, and causes a ‘misalignment’ of 11.3° between the C1′···C1′ vector (red dotted line) and the base-centered mean y-axis (horizontal line):
atan2(2, 10) * 180 / pi = 11.3°
To a first order approximation, that is the difference in twist angle. So whenever a wobble pair is next to a normal Watson-Crick pair, there would be a ~11° “observed” discrepancy with 3DNA calculated twist angle. Moreover, when a G–U wobble is next to a U–G wobble pair or vice versa, the difference would be doubled to ~22°.