G-quadruplexes (hereafter referred to as G4) are a common type of higher-order DNA and RNA structures formed from G-rich sequences. The building block of G4 is a tetrad of guanines in a cyclic planar alignment, with four G+G pairs (cW+M type, see Figure below). A G4 structure is formed by stacking of G-tetrads and stabilized by cations at the center of the layers. G4 structures are polymorphic: the four strands can be parallel or anti-parallel, and loops connecting them can be of different types: lateral (edgewise), diagonal, or propeller (double-chain reversal). Moreover, G4 structures can be intra- or intermolecular, and even contain bulges.
From its initial releases, DSSR was able to detect G-tetrads, and listed them in a separate section. As of v1.7.0-2017oct19, DSSR has integrated existing features and created a new module to automatically identify and fully characterize G4 structures. The underlying algorithms have been further refined in v1.7.1-2017nov01, which was tested against all nucleic-acid-containing structures in the PDB.
Characterizations of three representative G4 examples (PDB entries 2m4p, 2hy9, and 5hix) are shown below, illustrating salient features (e.g., different types of loops) automatically extracted by DSSR.
2m9p
stem#1[#1] layers=3 INTRA-molecular parallel bulged-strands=1
1 syn=---- WC-->Major area=8.38 rise=3.64 twist=33.34 nts=4 GGGG A.DG3,A.DG8,A.DG12,A.DG16
2 syn=---- WC-->Major area=10.73 rise=3.23 twist=32.42 nts=4 GGGG A.DG5,A.DG9,A.DG13,A.DG17
3 syn=---- WC-->Major nts=4 GGGG A.DG6,A.DG10,A.DG14,A.DG18
strand#1* +1 DNA syn=--- nts=3 GGG A.DG3,A.DG5,A.DG6 bulged-nts=1 T A.DT4
strand#2 +1 DNA syn=--- nts=3 GGG A.DG8,A.DG9,A.DG10
strand#3 +1 DNA syn=--- nts=3 GGG A.DG12,A.DG13,A.DG14
strand#4 +1 DNA syn=--- nts=3 GGG A.DG16,A.DG17,A.DG18
loop#1 type=propeller strands=[#1,#2] nts=1 T A.DT7
loop#2 type=propeller strands=[#2,#3] nts=1 T A.DT11
loop#3 type=propeller strands=[#3,#4] nts=1 T A.DT15
2hy9
stem#1[#1] layers=3 INTRA-molecular anti-parallel
1 syn=ss-s Major-->WC area=13.69 rise=3.14 twist=19.08 nts=4 GGGG 1.DG4,1.DG10,1.DG18,1.DG22
2 syn=--s- WC-->Major area=13.40 rise=3.05 twist=28.05 nts=4 GGGG 1.DG5,1.DG11,1.DG17,1.DG23
3 syn=--s- WC-->Major nts=4 GGGG 1.DG6,1.DG12,1.DG16,1.DG24
strand#1 +1 DNA syn=s-- nts=3 GGG 1.DG4,1.DG5,1.DG6
strand#2 +1 DNA syn=s-- nts=3 GGG 1.DG10,1.DG11,1.DG12
strand#3 -1 DNA syn=-ss nts=3 GGG 1.DG18,1.DG17,1.DG16
strand#4 +1 DNA syn=s-- nts=3 GGG 1.DG22,1.DG23,1.DG24
loop#1 type=propeller strands=[#1,#2] nts=3 TTA 1.DT7,1.DT8,1.DA9
loop#2 type=lateral strands=[#2,#3] nts=3 TTA 1.DT13,1.DT14,1.DA15
loop#3 type=lateral strands=[#3,#4] nts=3 TTA 1.DT19,1.DT20,1.DA21
5hix
stem#1[#1] layers=4 inter-molecular anti-parallel
1 syn=s--s Major-->WC area=12.93 rise=3.64 twist=16.82 nts=4 GGGG A.DG1,B.DG4,A.DG12,B.DG9
2 syn=-ss- WC-->Major area=18.96 rise=3.71 twist=35.87 nts=4 GGGG A.DG2,B.DG3,A.DG11,B.DG10
3 syn=s--s Major-->WC area=15.16 rise=3.64 twist=18.64 nts=4 GGGG A.DG3,B.DG2,A.DG10,B.DG11
4 syn=-ss- WC-->Major nts=4 GGGG A.DG4,B.DG1,A.DG9,B.DG12
strand#1 +1 DNA syn=s-s- nts=4 GGGG A.DG1,A.DG2,A.DG3,A.DG4
strand#2 -1 DNA syn=-s-s nts=4 GGGG B.DG4,B.DG3,B.DG2,B.DG1
strand#3 -1 DNA syn=-s-s nts=4 GGGG A.DG12,A.DG11,A.DG10,A.DG9
strand#4 +1 DNA syn=s-s- nts=4 GGGG B.DG9,B.DG10,B.DG11,B.DG12
loop#1 type=diagonal strands=[#1,#3] nts=4 TTTT A.DT5,A.DT6,A.DT7,A.DT8
loop#2 type=diagonal strands=[#2,#4] nts=4 TTTT B.DT5,B.DT6,B.DT7,B.DT8
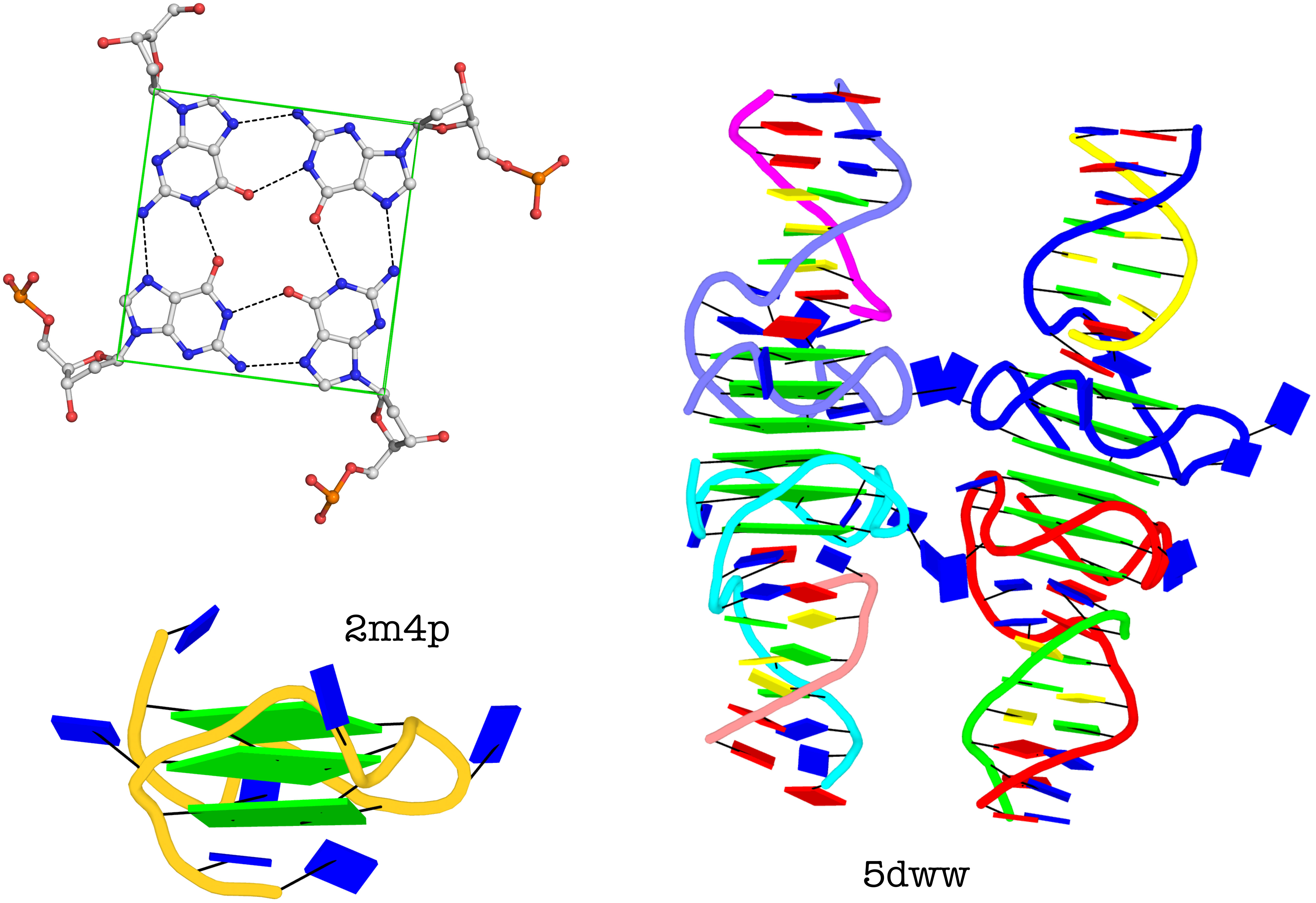
The molecular structure of the G-tetrad and two G4 structures in schematics representation. Upper left: atomic structure of G-tetrad, the building block of G4 structures. Here the green ‘square’ is created by connecting the C1’ atoms of the guanosines, and it is used to simplify the representation of G4 structures of PDB entries 2m4p (lower left) and 5dww (right). Note that the asymmetric unit of 5dww contains four biological units, which are coaxially stacked in two columns.
The DSSR output for PDB entry 5dww is listed below, showing the differences of a G4-helix vs. a G4-stem.
5dww
Note: a G4-helix is defined by stacking interactions of G4-tetrads, regardless
of backbone connectivity, and may contain more than one G4-stem.
helix#1[#2] layers=6 inter-molecular stems=[#1,#2]
1 syn=---- WC-->Major area=10.64 rise=3.54 twist=28.10 nts=4 GGGG A.DG3,A.DG7,A.DG11,A.DG16
2 syn=.--- WC-->Major area=11.63 rise=3.65 twist=31.14 nts=4 GGGG A.DG2,A.DG6,A.DG10,A.DG15
3 syn=---- WC-->Major area=28.36 rise=3.31 twist=-9.78 nts=4 GGGG A.DG1,A.DG5,A.DG9,A.DG14
4 syn=---- Major-->WC area=11.60 rise=3.75 twist=29.43 nts=4 GGGG C.DG1,C.DG14,C.DG9,C.DG5
5 syn=---- Major-->WC area=10.35 rise=3.49 twist=28.74 nts=4 GGGG C.DG2,C.DG15,C.DG10,C.DG6
6 syn=---- Major-->WC nts=4 GGGG C.DG3,C.DG16,C.DG11,C.DG7
strand#1 DNA syn=-.---- nts=6 GGGGGG A.DG3,A.DG2,A.DG1,C.DG1,C.DG2,C.DG3
strand#2 DNA syn=------ nts=6 GGGGGG A.DG7,A.DG6,A.DG5,C.DG14,C.DG15,C.DG16
strand#3 DNA syn=------ nts=6 GGGGGG A.DG11,A.DG10,A.DG9,C.DG9,C.DG10,C.DG11
strand#4 DNA syn=------ nts=6 GGGGGG A.DG16,A.DG15,A.DG14,C.DG5,C.DG6,C.DG7
......
List of 4 G4-stems
Note: a G4-stem is defined as a G4-helix with backbone connectivity.
Bulges are also allowed along each of the four strands.
stem#1[#1] layers=3 INTRA-molecular parallel
1 syn=---- WC-->Major area=11.63 rise=3.65 twist=31.14 nts=4 GGGG A.DG1,A.DG5,A.DG9,A.DG14
2 syn=.--- WC-->Major area=10.64 rise=3.54 twist=28.10 nts=4 GGGG A.DG2,A.DG6,A.DG10,A.DG15
3 syn=---- WC-->Major nts=4 GGGG A.DG3,A.DG7,A.DG11,A.DG16
strand#1 +1 DNA syn=-.- nts=3 GGG A.DG1,A.DG2,A.DG3
strand#2 +1 DNA syn=--- nts=3 GGG A.DG5,A.DG6,A.DG7
strand#3 +1 DNA syn=--- nts=3 GGG A.DG9,A.DG10,A.DG11
strand#4 +1 DNA syn=--- nts=3 GGG A.DG14,A.DG15,A.DG16
loop#1 type=propeller strands=[#1,#2] nts=1 T A.DT4
loop#2 type=propeller strands=[#2,#3] nts=1 T A.DT8
loop#3 type=propeller strands=[#3,#4] nts=2 TT A.DT12,A.DT13
--------------------------------------------------------------------------
stem#2[#1] layers=3 INTRA-molecular parallel
1 syn=---- WC-->Major area=11.60 rise=3.75 twist=29.43 nts=4 GGGG C.DG1,C.DG5,C.DG9,C.DG14
2 syn=---- WC-->Major area=10.35 rise=3.49 twist=28.74 nts=4 GGGG C.DG2,C.DG6,C.DG10,C.DG15
3 syn=---- WC-->Major nts=4 GGGG C.DG3,C.DG7,C.DG11,C.DG16
strand#1 +1 DNA syn=--- nts=3 GGG C.DG1,C.DG2,C.DG3
strand#2 +1 DNA syn=--- nts=3 GGG C.DG5,C.DG6,C.DG7
strand#3 +1 DNA syn=--- nts=3 GGG C.DG9,C.DG10,C.DG11
strand#4 +1 DNA syn=--- nts=3 GGG C.DG14,C.DG15,C.DG16
loop#1 type=propeller strands=[#1,#2] nts=1 T C.DT4
loop#2 type=propeller strands=[#2,#3] nts=1 T C.DT8
loop#3 type=propeller strands=[#3,#4] nts=2 TT C.DT12,C.DT13