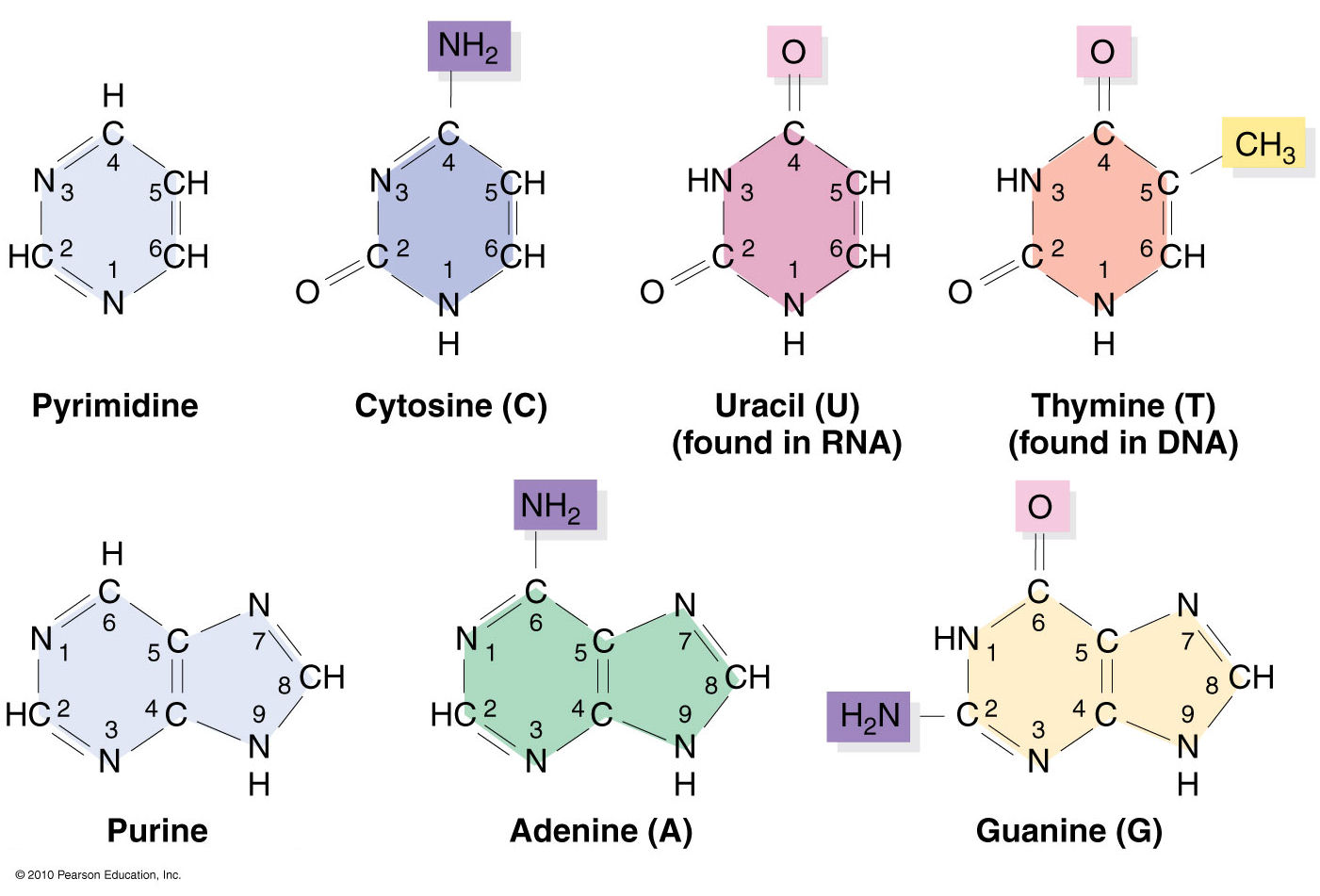
Nucleic acids structural bioinformatics starts with the identification of nucleotides (nts) from atomic coordinates. As biopolymers, RNA and DNA have standard IUPAC names of atoms for the five bases (see the Figure below), sugars (ending with prime, e.g., C1’, O2’), and the phosphate (P, OP1, and OP2). The atomic coordinates (in PDB or mmCIF format) from the Protein Data Bank (PDB) follow the convention.
Trained as a chemist, I am aware that the bases are aromatic, heterocyclic compounds (purines and pyrimidines). Moreover, the five standard bases (A, C, G, T, and U) also share a six-membered ring, with atoms named consecutively (N1, C2, N3, C4, C5, C6). This special feature can be employed to identify nts automatically, from PDB atomic coordinates. The ring skeleton is not influenced by protonation states, tautomeric forms, or modifications in base, sugar or phosphate. Early versions of 3DNA (up to v2.0) used only N1, C2, and C6 atoms to identify an nt: an additional N9 as purine, otherwise as pyrimidine. In 3DNA v2.3 and DSSR, the procedure has been refined to take advantage of all available rings atoms. It is thus more robust against distortions and still works even when any of the N1, C2, C6, or N9 atoms are mutated or missing. This blog post provides further technical details on how the method works.
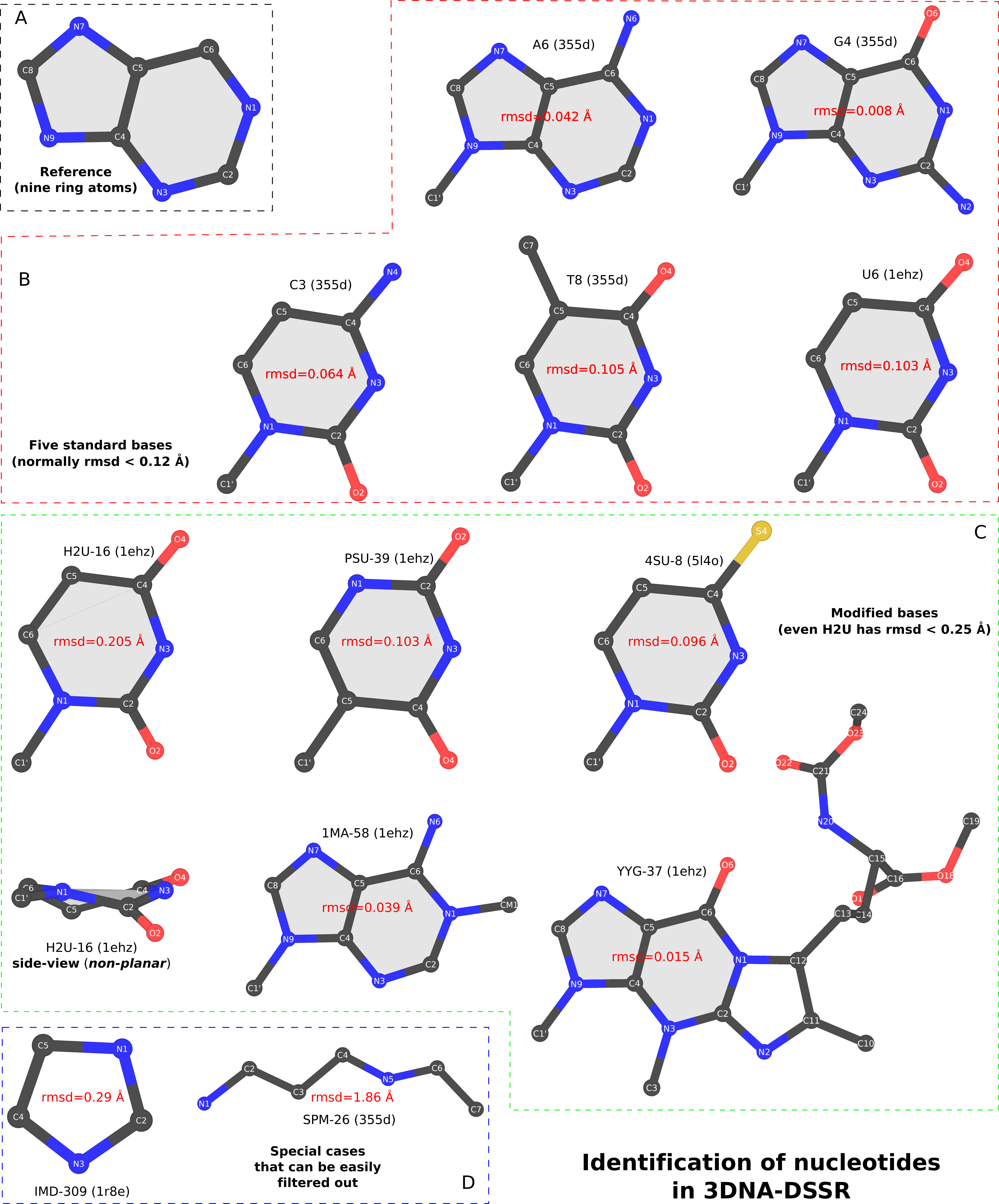
The template used to identify nts is a purine, with nine base ring atoms. Purine is chosen since it contains atoms of the six-membered ring and N7, C8, and N9. Its atomic coordinates in PDB format are shown below. The coordinates are taken from ‘G’ in the standard reference frame ($X3DNA/config/Atomic_G.pdb). Using ‘A’ as reference won’t make any difference since the RMSD between them is only 0.038 Å.
ATOM 1 N9 G A 1 -1.289 4.551 0.000 1.00 0.00 N ATOM 2 C8 G A 1 0.023 4.962 0.000 1.00 0.00 C ATOM 3 N7 G A 1 0.870 3.969 0.000 1.00 0.00 N ATOM 4 C5 G A 1 0.071 2.833 0.000 1.00 0.00 C ATOM 5 C6 G A 1 0.424 1.460 0.000 1.00 0.00 C ATOM 6 N1 G A 1 -0.700 0.641 0.000 1.00 0.00 N ATOM 7 C2 G A 1 -1.999 1.087 0.000 1.00 0.00 C ATOM 8 N3 G A 1 -2.342 2.364 0.001 1.00 0.00 N ATOM 9 C4 G A 1 -1.265 3.177 0.000 1.00 0.00 C
The nt-identification process begins with a mapping of at least three atoms in the purine, followed by a least-squares fit between corresponding atoms. For the five standard bases and most modified ones, the RMSD is normally less than 0.12 Å, as seen in the Figure below. Even the unsaturated dihydrouridine in tRNA has an RMSD of less than 0.25 Å: for the yeast phenylalanine tRNA (PDB id: e1ehz), for example, it is 0.205 Å for H2U-16, and 0.226 Å for H2U-17. DSSR uses a cutoff of 0.28 Å, covering essentially all nucleotides in the PDB. As an extreme case, the DA1 residue on chain T of PDB id 4ki4 has only three base atoms: N7, C8, and N9 (i.e., no atoms from the six-membered ring). With an RMSD of only 0.005 Å, DSSR still takes it as an nt, properly assigned as ‘A’.
Molecular dynamics (MD) simulations sometimes produce heavily distorted bases, which is over the default cutoff. Users may change the cutoff to a larger value to accommodate such unusual cases.
In addition to dihydrouridine, the above Figure also shows pseudouridine (PSU), 1-methyladenosine (1MA), 4-thiouridine (4SU), and the heavily modified YYG in tRNA. They are all easily identified using the same scheme. Since the nt-identification method concentrates on base rings, modifications in sugar or the phosphate group do not pose any problem. For example, in tRNA 1ehz, DSSR also identifies O2’-methylguanosine (OMG) and O2’-methylcytidine (OMC) as modified nts.
Two special cases worth mentioning. The ligand IMD in PDB id 1r8e has a five-membered ring. Its atoms are named similarly to those of an nt, and the fitted RMSD is only 0.29 Å. IMD can be filtered out by its missing of the C6 atom and having an N1—C5 covalent bond. The ligand SPM in PDB id 355d is a linear molecule, and its RMSD (1.86 Å) is clearly far off to be taken as an nt.
Another particular case (of a different kind) is the abasic sites, especially in X-ray crystal structures in the PDB. By definition, abasic sites do not have base atoms available. Thus the described method is not applicable to their characterization as nts. As of v1.7.3-2017dec26, however, DSSR has also incorporated abasic sites into the analysis pipeline, by default. The program checks backbone linkage and residue name for appropriate nt assignment. The abasic sites could constitute part of (internal) loops which would otherwise be broken into segments by DSSR.
Overall, I feel confident to say that 3DNA-DSSR has practically solved the problem of identifying nts from atomic coordinates. The method detailed herein (and outlined in the DSSR paper) is simple and easy to understand/implement. Moreover, it has been extensively tested in real-world applications for well over a decade. I’ve yet to find a single case where it does not work as expected.