The paper, titled DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL, has just been published in Nucleic Acids Research (online on May 22, 2020). Here is the abstract:
Sophisticated analysis and simplified visualization are crucial for understanding complicated structures of biomacromolecules. DSSR (Dissecting the Spatial Structure of RNA) is an integrated computational tool that has streamlined the analysis and annotation of 3D nucleic acid structures. The program creates schematic block representations in diverse styles that can be seamlessly integrated into PyMOL and complement its other popular visualization options. In addition to portraying individual base blocks, DSSR can draw Watson-Crick pairs as long blocks and highlight the minor-groove edges. Notably, DSSR can dramatically simplify the depiction of G-quadruplexes by automatically detecting G-tetrads and treating them as large square blocks. The DSSR-enabled innovative schematics with PyMOL are aesthetically pleasing and highly informative: the base identity, pairing geometry, stacking interactions, double-helical stems, and G-quadruplexes are immediately obvious. These features can be accessed via four interfaces: the command-line interface, the DSSR plugin for PyMOL, the web application, and the web application programming interface. The supplemental PDF serves as a practical guide, with complete and reproducible examples. Thus, even beginners or occasional users can get started quickly, especially via the web application at http://skmatic.x3dna.org.
A brief history on DNA/RNA schematics as implemented in SCHNAaP/SCHNArP, 3DNA, and now in DSSR:
The idea of representing bases and WC-pairs as rectangular blocks came from the pioneering work of Calladine et al. (27,28) The block schematics were first implemented in the pair of SCHNAaP/SCHNArP programs (29,30) for rigorous analysis and reversible rebuilding of double-helical nucleic acid structures. The algorithms that underpinned SCHNAaP/SCHNArP laid the foundation of ‘analyze’ and ‘rebuild’, two core components of the 3DNA suite of programs (31–33). 3DNA also takes advantage of the standard base reference frame (34), and comprises quite a few other related programs. One of them is ‘blocview’, a script which calls several 3DNA utility programs to generate individual base blocks and set the view, MolScript (35) to produce backbone ribbons, and Raster3D (36) to render the composite image. The 3DNA ‘blocview’ schematics catch characteristic attributes of nucleic acid structures. They have gradually become popular and been adopted into the RCSB PDB (1) and the NDB (37), and then propagated into other bioinformatics resources (e.g., the ‘RNA Structure Atlas’ website hosted by the Leontis-Zirbel RNA group).
DSSR supersedes ‘blocview’ by eliminating all the internal and external dependencies of the 3DNA utility program. DSSR produces block representations, not only of individual bases but also WC-pairs and G-tetrads, that can be fed directly into PyMOL. The DSSR-PyMOL integration is easier to use, has more features, and produces better schematics than the original 3DNA-blocview approach.
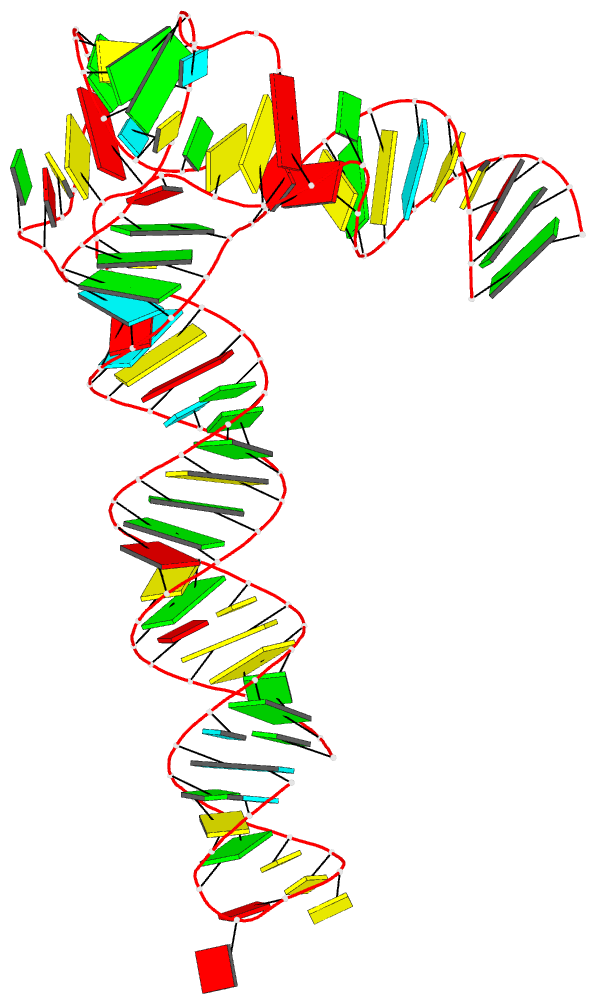 |
 |
| Schematic image for PDB entry 6ol3 auto-generated via the DSSR-PyMOL integration | Cover image of the May 2020 issue of the RNA Journal, “generated using 3DNA/blocview and PyMol software by the Nucleic Acid Database” |
Indeed, the base block schematics have continuously evolved for over two decades, as appreciated in the acknowledgements:
I would like to thank Christopher A. Hunter, Christopher R. Calladine, Helen M. Berman, Catherine L. Lawson, Zukang Feng, Wilma K. Olson and Harmen J. Bussemaker for their helpful input on the block schematic during its continuous evolution for over two decades. I appreciate Thomas Holder (PyMOL Principal Developer, Schrödinger, Inc.) for writing the DSSR plugin for PyMOL, and for providing insightful comments on the manuscript and the web application interface. I also thank Jessalyn Lu and Yin Yin Lu for proofreading the manuscript, and the user community for feedback.
Notably, the supplemental PDF has been diligently written to serve as a practical guide, with complete and reproducible examples. In fact, the paper concludes with the following two sentences:
Finally, all results reported here are completely reproduceable (see the supplemental PDF). Any questions related to this work are welcome and will be openly addressed on the 3DNA Forum (http://forum.x3dna.org).
