The Kribelbauer et al. article, Towards a mechanistic understanding of DNA methylation readout by transcription factors has recently been published in the Journal of Molecular Biology (JMB). I am honored to be among the author list, and I learned a lot during the process. For the project, I added the --methyl-C (short-form: --5mc) option to SNAP (v1.0.6-2019sep30) for the automatic identification and annotation of DNA-transcription factor (TF) complexes containing 5-methyl-cytosine (5mC). The results are presented in a dynamic table, easily accessible at URL http://snap-5mc.x3dna.org, and summarized in Fig. 1 “Structural basis of how TFs recognize methylated DNA” (see below) of the JMB paper.
Details on the SNAP-enabled curation of TF-DNA complexes containing 5mC from atomic coordinates in the Protein Data Bank (PDB) are available in a tutorial page at http://snap-5mc.x3dna.org/tutorial. In essence, the process can be easily understood via a concrete example with PDB id 4m9e, as shown below.
x3dna-snap --methyl-C --type=base -i=4m9e.pdb -o=4m9e-5mC.out
Here the --methyl-C option is specific for 5mC-DNA, and --type=base ensures that at least one DNA base atom is contacting protein amino acid(s). If these conditions are fulfilled, SNAP would produce two additional 5mC-related files, apart from the normal output file (i.e., 4m9e-5mC.out, as specified in the example):
- 4m9e-5mC.txt — a simple text file with the following contents:
4m9e:B.5CM5: stacking-with-A.ARG443 is-WC-paired is-in-duplex [+]:GcG/cGC 4m9e:C.5CM5: other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG
- 4m9e-5mC.pdb — a corresponding PDB file, potentially multi-model, two as in this case. Moreover, the cluster of interacting residues (DNA nucleotides and protein amino acids) is oriented in the standard base reference frame of 5mC, allowing for easy comparison and direct overlap of multiple clusters.
In practice, SNAP needs to take care of many details for the automatic identification and annotation of 5mC-DNA-TF complexes directly from PDB entries. For example, 5mC in DNA is designated 5CM and the 5-methyl carbon atom is named C5A in the PDB (see the blogpost 5CM and 5MC, two forms of 5-methylcytosine in the PDB). Moreover, the --type=base option is employed to ensure that base atoms (regardless sugar-phosphate atoms) of 5mC are directly involved in interactions with amino acids.
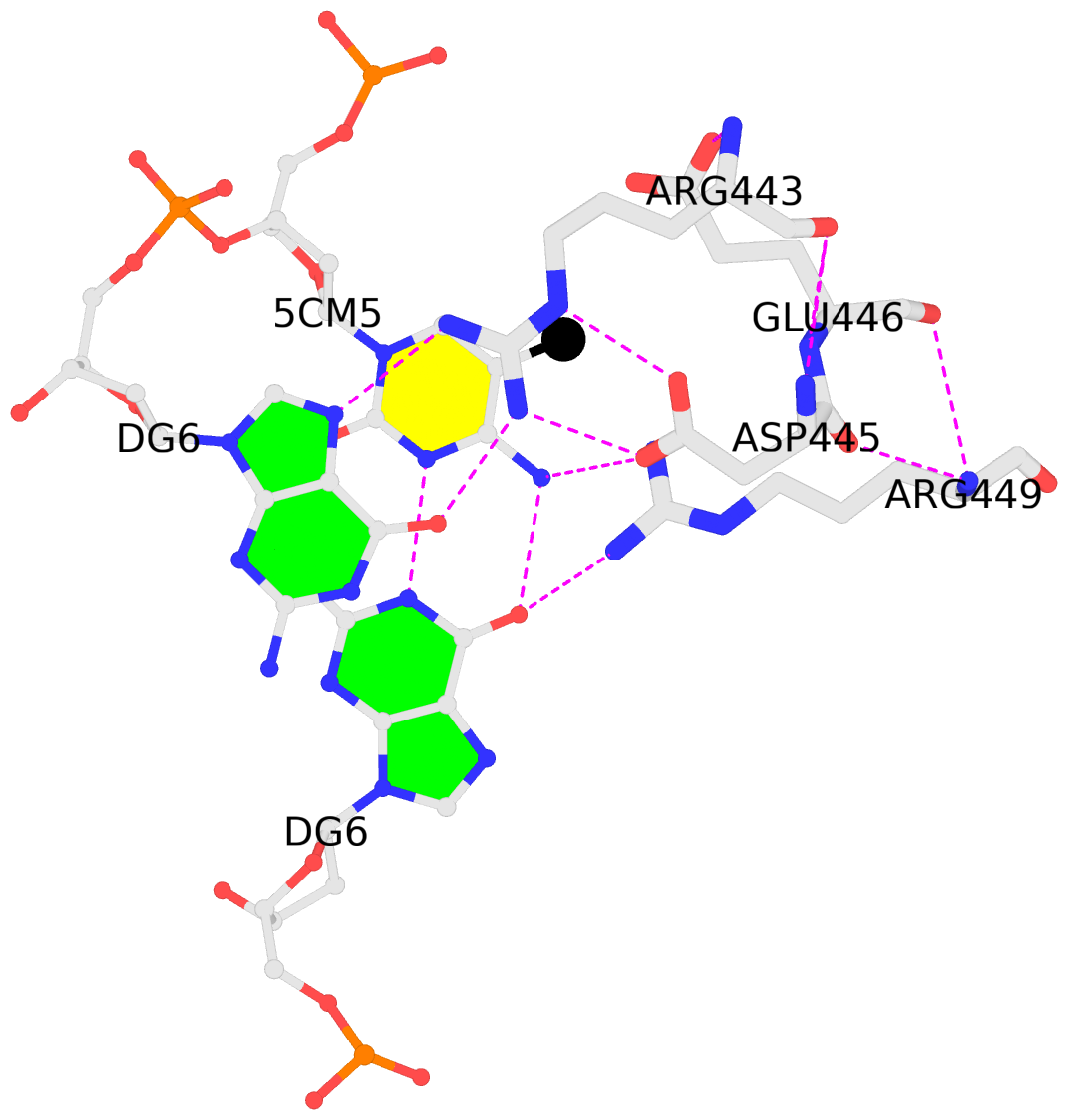
It is also worth noting the combined use of DSSR for the generation of molecular images (rendered with PyMOL), as shown below. Here the DSSR options --block-file=fill-hbond (fill to fill base rings and hbond to draw hydrogen bonds) and --cartoon-block=sticks-label are used. The 3DNA DSSR/SNAP combo is a unique and powerful toolset for structural bioinformatics, as demonstrated in DNAproDB from the Rohs lab (see my blogpost SNAP and DSSR in DNAproDB). The JMB paper represents yet another example. I can only expect to see more combined DSSR/SNAP applications in the future.