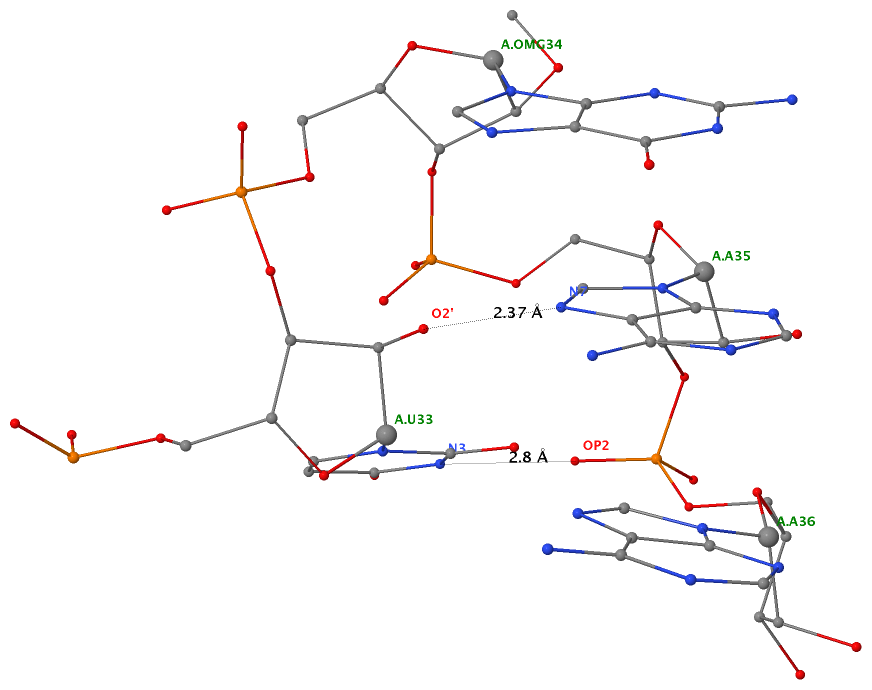
As of beta-r20-on-20130830, DSSR is able to detect two types of U-turns (see the figure below), the UNR-type (left) originally identified by Quigley and Rich [1976] in yeast phenylalanine tRNA, and the GNRA-type (right) later on established by Jucker and Pardi [1995] in GNRA tetra loops. See the Gutell et al. paper Predicting U-turns in Ribosomal RNA with Comparative Sequence Analysis for a more extensive account of U-turns.
As its name implies, a U-turn is characterized by a reversal of the RNA backbone direction within a few nucleotides. Among other factors, the U-turn is stabilized by two key H-bonding interactions, illustrated in dotted lines in the figure below.
 |
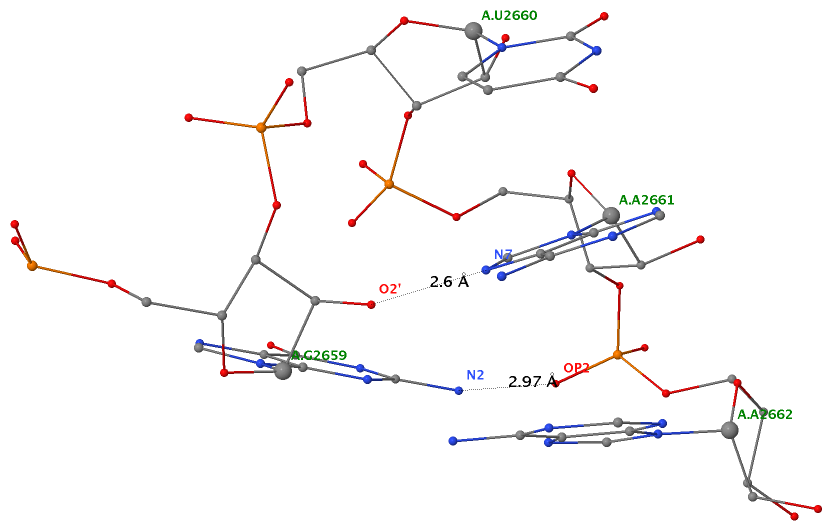 |
| UNR-type (1ehz) | GNRA-type (1msy) |
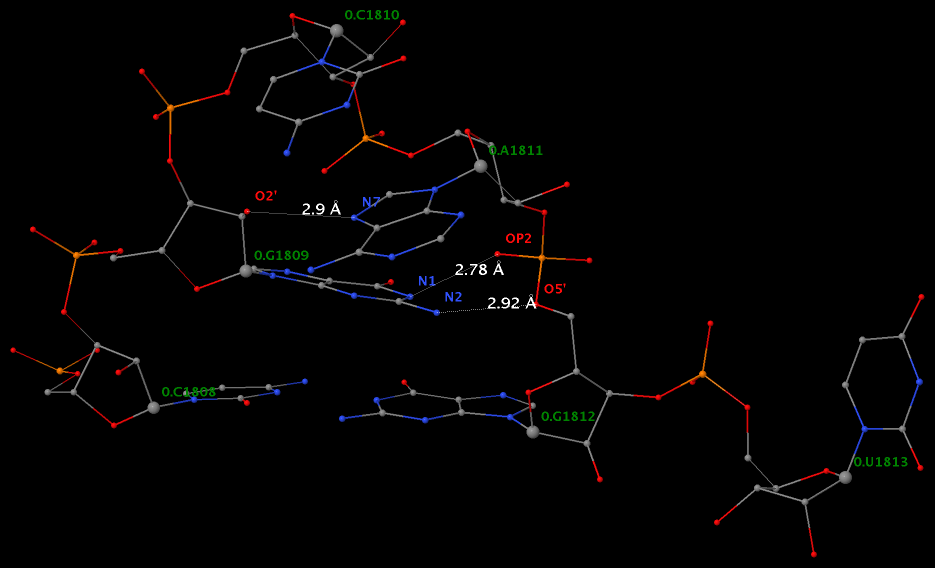
Applying DSSR to 1jj2 (the crystal structure of the Haloarcula marismortui large ribosomal subunit) led to the identification of over 30 cases. In addition to the well-documented UNR- and GNRA-type U-turns, the program also finds other variants. An example is shown below, where the U-turn is within a GCA triloop instead of a GNRA tetraloop. Here, the N1 (not N2) atom of G1809 forms an H-bond with OP2 of G1812. The G1809 N2 atom is H-bonded to G1812 O5′ to further stabilize the U-turn.
An examination of the chemical structure of the nitrogenous bases (see figure below) shows clearly other possibilities to connect RNA base donors to the phosphate oxygen acceptors. DSSR allows for the exploration of such variations, and more.