Recently, I (together with Drs. Wilma Olson and Harmen Bussemaker – a team with a unique combination of complementary expertise) published a new article in Nucleic Acids Research (NAR): The RNA backbone plays a crucial role in mediating the intrinsic stability of the GpU dinucleotide platform and the GpUpA/GpA mini duplex. The key findings of this work are summarized in the abstract:
The side-by-side interactions of nucleobases contribute to the organization of RNA, forming the planar building blocks of helices and mediating chain folding. Dinucleotide platforms, formed by side-by-side pairing of adjacent bases, frequently anchor helices against loops. Surprisingly, GpU steps account for over half of the dinucleotide platforms observed in RNA-containing structures. Why GpU should stand out from other dinucleotides in this respect is not clear from the single well-characterized H-bond found between the guanine N2 and the uracil O4 groups. Here, we describe how an RNA-specific H-bond between O2’(G) and O2P(U) adds to the stability of the GpU platform. Moreover, we show how this pair of oxygen atoms forms an out-of-plane backbone ‘edge’ that is specifically recognized by a non-adjacent guanine in over 90% of the cases, leading to the formation of an asymmetric miniduplex consisting of ‘complementary’ GpUpA and GpA subunits. Together, these five nucleotides constitute the conserved core of the well-known loop-E motif. The backbone-mediated intrinsic stabilities of the GpU dinucleotide platform and the GpUpA/GpA miniduplex plausibly underlie observed evolutionary constraints on base identity. We propose that they may also provide a reason for the extreme conservation of GpU observed at most 5’-splice sites.
As a nice surprise, this publication was selected by NAR as a featured article! According to the NAR website:
Featured Articles highlight the best papers published in NAR. These articles are chosen by the Executive Editors on the recommendation of Editorial Board Members and Referees. They represent the top 5% of papers in terms of originality, significance and scientific excellence.
I feel very gratified with the “extra” recognition. From my own perspective, I can easily rank this paper as the top one in my publication list: from the very beginning, I has been struck by the simplicity and elegance of the GpU story. Hopefully, time will verify the validity of this scientific contribution.
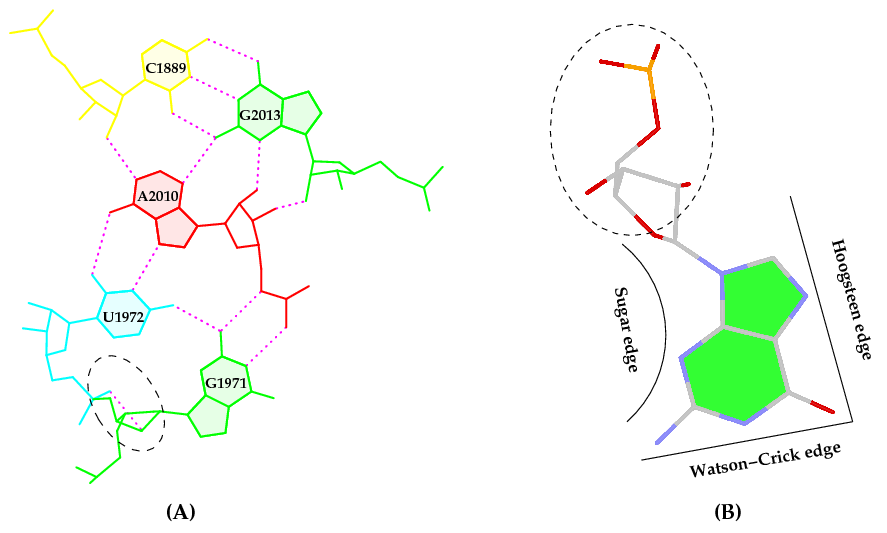
Behind the hood, though, there is a long, complex (sometimes perplexing), yet interesting story associated with this work. Here is how it got started. While writing the 3DNA 2008 Nature Protocols (NP) paper, I selected the (previously undocumented) ‘-p’ option of find_pair to showcase its capability to identify higher-order base associations, using the large ribosomal subunit (1jj2) as an example. I noticed the unexpected O2’(G)⋅⋅⋅O2P H-bond within the GpU dinucleotide platform in a pentaplet (Figure A below). I was/am well aware of Leontis-Westholf’s pioneering work on Geometric nomenclature and classification of RNA base pairs which involves three distinct edges – the Watson-Crick edge, the Hoogsteen edge, and the Sugar edge, yet without taking into consideration of possible sugar-phosphate backbone interactions (Figure B below). So I decided to double-check, just to be sure that the H-bond was not spurious due to defects in the H-bond detecting scheme of find_pair, and the finding was very surprising.
The following section was re-added into the 3DNA NP paper in the very last revision:
It is also worth noting that the G1971–U1972 platform is stabilized not only by the well-characterized G(N2)⋅⋅⋅U(O4) H-bond interaction, but also by a little-noticed G(O2’)⋅⋅⋅U(O2P) sugar-phosphate backbone interaction (Fig. 6a). Examination of the 50S large ribosomal unit (1JJ2) alone reveals ten such double H-bonded G–U platforms, far more occurrences than those registered by any other dinucleotide platform (including A–A) in this structure. Apparently, the G–U platform is more stable than other platforms with only a single base–base H-bond interaction. We are currently investigating this overrepresented G–U dinucleotide platform in other RNA structures. (p.1226)