Recently I was a bit surprised to find that the methyl group is named differently in the PDB: C7 in DT8 (thymine) of B-DNA 355d, CM5 in 5MC40 (5-methylated C) of tRNA 1ehz, and C5M in 5MU54 (5-methylated U, i.e., T) of the same tRNA 1ehz. See the three figures below for details.
I know that the previously named C5M of thymine in DNA has been renamed C7 as a result of the 2007 remediation effort (PDB v3). However, browsing through the wwPDB Remediation website and reading carefully the article Remediation of the protein data bank archive, I failed to see explanations of the obvious inconsistency of CM5 (5MC40) vs C5M (5MU54) in the nomenclature of the 5-methyl group in the same tRNA entry 1ehz, except for the following note:
As with the Chemical Component Dictionary, names for standard amino acids and nucleotides follow IUPAC recommendations (10) with the exception of the well-established convention for C-terminal atoms OXT and HXT. These nomenclature changes have been applied to standard polymeric chemical components only.
5-methyl is named C7 in DT8 of the DNA entry 355d
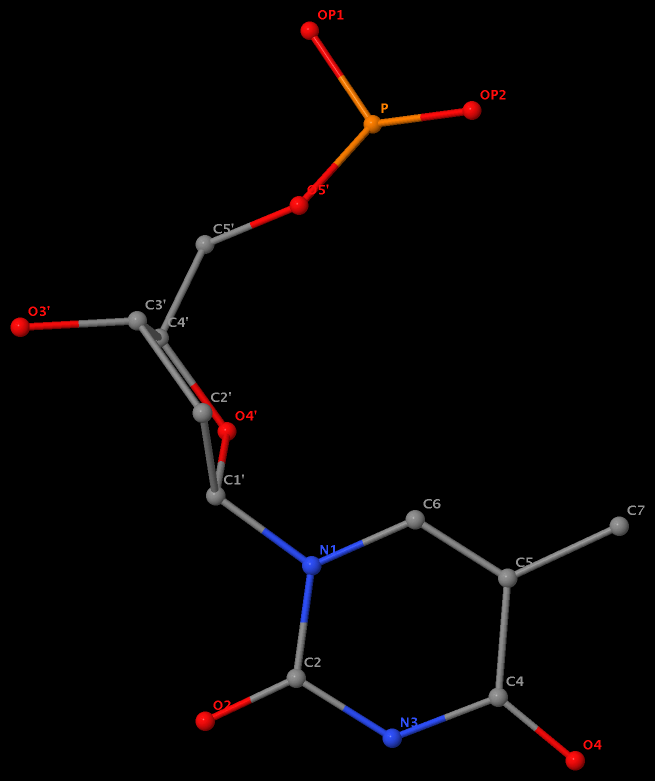
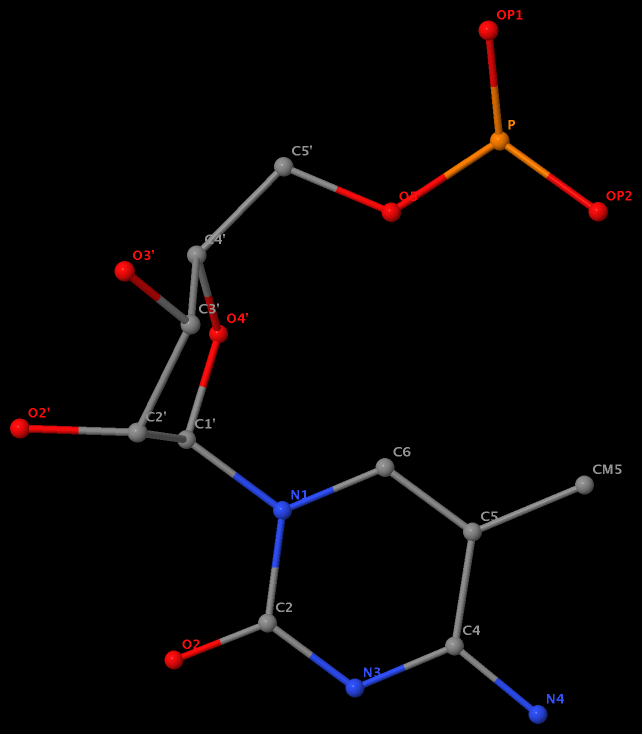
5-methyl is named CM5 in 5MC40 of the RNA entry 1ehz
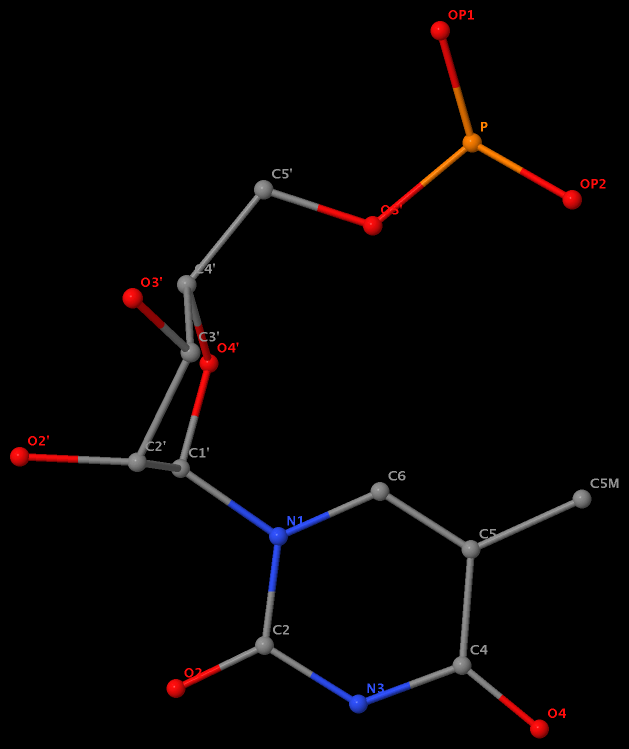
5-methyl is named C5M in 5MU54 of the RNA entry 1ehz
Am I missing something obvious? If you have any further information, please leave a comment. Whatever the case, it helps (at least won’t hurt) to know the naming discrepancy for those who care about the small methyl group in nucleic acid structures.

Recently, I upgraded my local ViennaRNA package installation from v2.0.7 to v2.1.3 on my Mac. Following Quickstart in the INSTALL file, I ran ./configure successfully, but make aborted with error messages. Since I previously had a working copy of the software, it must be configuration issues when I compiled this new version. After a few iterations of checking the error message and reading through the INSTALL file, I came up with the following settings:
./configure --disable-openmp --without-perl
make
sudo make install
Apart from some warning messages, the above make command ran successfully.
This post serves mainly as a note for my own reference. Hopefully, the information may prove useful to others who try to install the versatile ViennaRNA package on a Mac OS X machine.

I’ve come up with a preliminary web-interface to DSSR, currently accessible at URL http://web.x3dna.org/dssr. The DSSR web-interface has been tested on Safari, Firefox, Chrome, and IE, with satisfying results. A screenshot of the home page is given below, using 1msy as an example:

After clicking the Submit button, users will be presented with the result page of a DSSR run. The beginning portion of the above example is as follows:

Note that the DSSR web-interface is being provided via a shared web hosting service, thus it has limited resources. Specifically, the uploaded file cannot be larger than two megabytes (2MB), and the process could be slow. Additionally, the file must have an extension of .pdb or .cif. To take full advantage of what DSSR has to offer, please install and run the software locally.
By design, DSSR is self-contained, command-line driven, with zero dependance on third-party libraries. Such features make it straightforward to build a GUI- or web-interface to DSSR, or integrate the program into other structural bioinformatics tools. As the need arises, I will refine the DSSR web-interface to better serve the community. The current simple, yet exploratory, web interface should make DSSR accessible to a much wider audience.

As of June 24, 2013, the number of 3DNA Forum registrations has passed the 1000 mark. On September 16, 2012, I wrote the post The number of 3DNA forum registrations has reached 500. Thus, in slightly over 9 months, the number has doubled, with approximately 2 registrations per day.
I am glad to see the steady increase of the 3DNA user base. Over the time, I have strived to be responsive to user questions, and made every effort to keep the forum spam free. By and large, employing simple 3DNA-related questions has turned out to be an effective anti-spam strategy. Since the launch of the new forum.x3dna.org in March 2012, I’ve received less than five requests (to the best of my memory) asking for help on registrations. As a recently example, a potential user got stuck with the question about what ‘w’ means in w3DNA. Based on user feedback, I have added hints to some questions to make their answers more obvious. Whatever the reasons, each reported issue has been promptly resolved.
With the release of DSSR and the continuous support of an enthusiastic user community, I have every reason to believe that 3DNA will gain more popularity in the years to come.

A new paper titled Analyzing and Building Nucleic Acid Structures with 3DNA has been published in JoVE (Journal of Visualized Experiments). Specifically, the article illustrates 3DNA’s unique capability to characterize and modify DNA structures at the level of the constituent base-pair steps, and highlights a new feature in v2.1 to analyze and align an ensemble of related structures determined with NMR or generated by MD simulations.
Here is the abstract:
The 3DNA software package is a popular and versatile bioinformatics tool with capabilities to analyze, construct, and visualize three-dimensional nucleic acid structures. This article presents detailed protocols for a subset of new and popular features available in 3DNA, applicable to both individual structures and ensembles of related structures. Protocol 1 lists the set of instructions needed to download and install the software. This is followed, in Protocol 2, by the analysis of a nucleic acid structure, including the assignment of base pairs and the determination of rigid-body parameters that describe the structure and, in Protocol 3, by a description of the reconstruction of an atomic model of a structure from its rigid-body parameters. The most recent version of 3DNA, version 2.1, has new features for the analysis and manipulation of ensembles of structures, such as those deduced from nuclear magnetic resonance (NMR) measurements and molecular dynamic (MD) simulations; these features are presented in Protocols 4 and 5. In addition to the 3DNA stand-alone software package, the w3DNA web server, located at http://w3dna.rutgers.edu, provides a user-friendly interface to selected features of the software. Protocol 6 demonstrates a novel feature of the site for building models of long DNA molecules decorated with bound proteins at user-specified locations.
A new section dedicated to the JoVE paper will be set up on the 3DNA Forum soon. It will contain all the data files and scripts so our published results can be strictly reproduced. The section should also serve as a platform for open discussions of related protocols.

Early on when I started on DNA structures, I read Saenger’s book Principles of Nucleic Acid Structure and became familiar with his classification of the 28 possible base-pairs (bps) for A, G, U(T), and C involving at least two (cyclic) hydrogen bonds (see figure below).

Later on, I read from the 2nd edition of The RNA World book a list of 29 bps compiled by Burkard, Turner & Tinoco. While the one bp discrepancy (28 vs 29) has been in my mind for quite a long while, I had never paid much attention to the issue until recently while adding classifications of RNA bps (among many other functionalities) to 3DNA. A Google search did not help solve the puzzle, so I decided to dig it out by comparing the two lists.
The Burkard et al. list is titled Structures of Base Pairs Involving at Least Two Hydrogen Bonds and it mentions specifically Saenger’s list:
The structures of 29 possible base pairs that involve at least two hydrogen bonds are given in Figures 1–5 (for further descriptions, see Saenger, in Principles of nucleic acid structure, p. 120. Springer-Verlag [1984]).
However, in the five figures, Burkard et al. do not provide the corresponding Saenger numbers (I to XXVIII, 1—28) for the 28 common bps; thus it is not immediately obvious which one (i.e., the new addition by Burkard et al.) is missing from Saenger’s list. Under careful scrutiny, the absent bp turns out to be the “G•C N3-amino, amino-N3” pair in Figure 3: “Six possible flipped purine-pyrimidine mismatches.” One example of such G+C pair is found in the 5S ribosomal RNA (chain 9, G3022—C3026) of Haloarcula marismortui in PDB entry 1vq8.
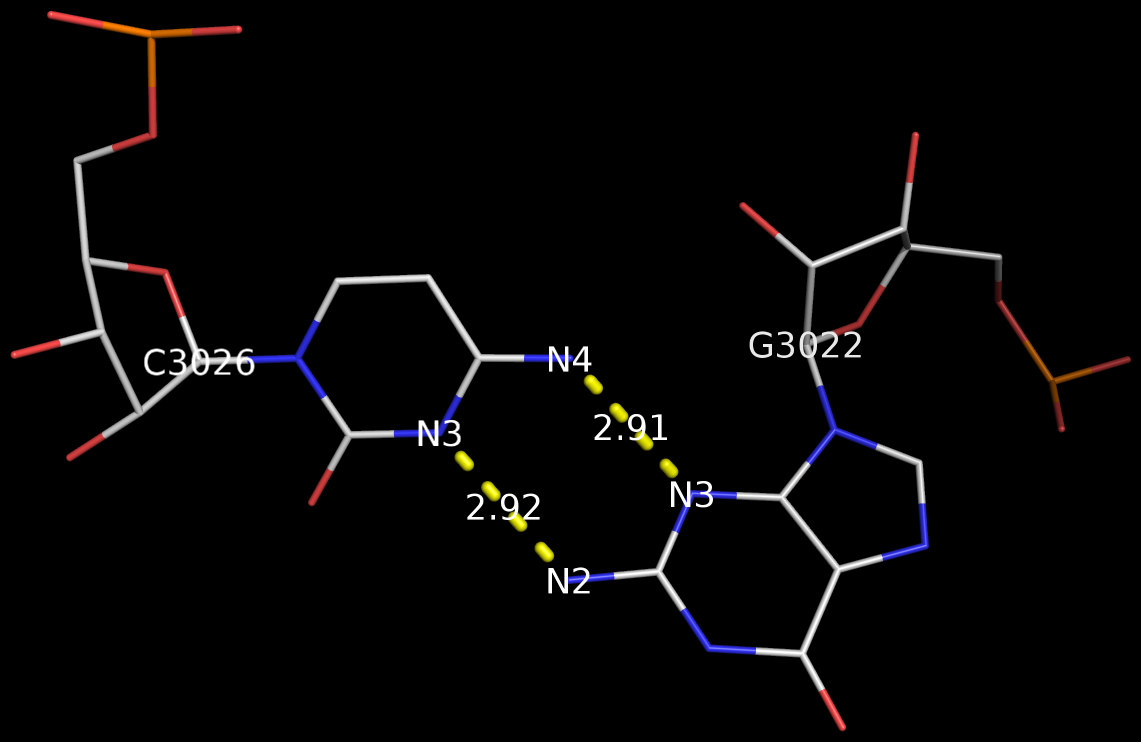
The above figure shows clearly that the G+C bp does indeed have two canonical H-bonds between base atoms, and it is difficult to speculate how it escaped Saenger’s selection criteria. In the upcoming new 3DNA component, I am listing this bp as number XXIX (29), along with the other 28 base pairs.

Recently, I came across the so-called Calcutta U-U base pair (bp) [see figure below] while reading articles on C-H…O contacts in nucleic acid structures. Not familiar with this named pair before, I was curious to find out what it’s about. After some searching, I traced the origin of the Calcutta U-U bp to the following two papers published by Sundaralingam’s group during the middle 1990s:
We have called the novel U•U base pair, where the Hoogsteen face of one of the pyrimidines is involved in a C5-H—O4 hydrogen bond, the ‘Calcutta Base Pair’, since it was announced at the International Seminar-cum-School on Macromolecular Crystallographic Data held in Calcutta, November 16-20, 1995.
We recently discovered a novel U•U base pair, referred to as the Calcutta base pair, in the crystal structure of an RNA hexamer UUCGCG (Ref. 18). The two uracil bases form a conventional N(3)-H…O(4) and an unconventional C(5)-H…O(2) hydrogen bond (Fig. 3a). The C-H…O interaction is entirely ‘voluntary’ and not ‘forced’, underlining its importance in base mispairing.
3DNA has no problem to identify the Calcutta U-U bps (or any pair for that matter); an example is shown below based on the RNA hexamer UUCGCG structure (PDB entry: 1osu) solved by Sundaralingam and colleagues.
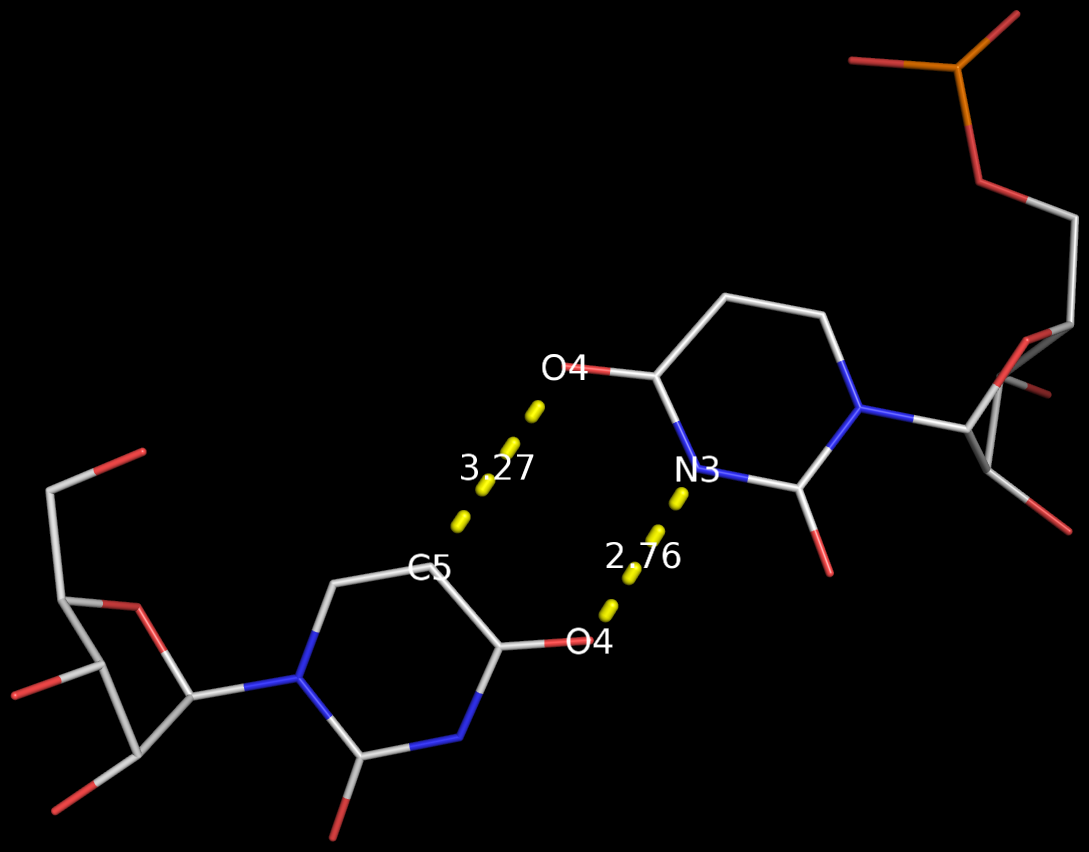
In the new 3DNA component I’ve been working on (and to be released soon), the Calcutta U-U pair is characterized as below:
1/A.U1 3/A.U2 [U-U] Calcutta 00-n/a tHW -MW
anti C3'-endo 8.9 --- anti C3'-endo 30.3
dcc=11.18 dnn=8.48 dmm=7.58 tor=-174.1
H-bonds[2]: "O4(carbonyl)-N3(imino)[2.76]; C5-O4(carbonyl)[3.27]"
Shear=-3.67 Stretch=-0.52 Stagger=-0.89
Buckle=-1.41 Propeller=-16.03 Opening=-90.67
The Calcutta pair is explicitly named, along with other named base pairs (e.g., Watson-Crick [WC], Wobble, and Hoogsteen bps). It is classified as type tHW (trans with Hoogsteen/WC interacting edges), following the commonly used Leontis-Westhof nomenclature. It does not belong to any of the 28 bps (00-n/a) with at least two conventional H-bonds, as categorized by Saenger. In 3DNA, the Calcutta U-U pair is of M-N type, designated as -MW.
Among the well-known named base pairs, some are after the scientists who discovered them (e.g., WC and Hoogsteen bps), while others are based on chemical/geometrical features (e.g., Wobble and Sheared G-A bps), or a combination of both (e.g., reversed WC/Hoogsteen bps). The Calcutta U-U pair is unique in that it is named after a place in India:
Kolkata, or Calcutta, is the capital of the Indian state of West Bengal. … While the city’s name has always been pronounced Kolkata or Kolikata in Bengali, the anglicized form Calcutta was the official name until 2001, when it was changed to Kolkata in order to match Bengali pronunciation.

Prior to v2.1, 3DNA does not provide any direct support for the analysis of molecular dynamics (MD) simulations trajectories of nucleic acid structures. Nevertheless, over the years, I noticed some significant applications of 3DNA in the active MD field; see my blog post (December 6, 2009) titled 3DNA in the PCCP nucleic acid simulations themed issue. In January 2011, I released a set of two Ruby scripts specifically aimed to facilitate the analysis of MD simulations trajectories. Thereafter (as of 3DNA v2.1), I have significantly refined and expanded the Ruby scripts, and consolidated the functionality under one umbrella, x3dna_ensemble with multiple sub-commands (analyze, block_image, extract, and reorient). I believe x3dna_ensemble would make it straightforward to analyze ensembles (NMR or MD simulations trajectories) of nucleic acid structures.
Under this background, I am glad to read recently an article titled Structure, Stiffness and Substates of the Dickerson-Drew Dodecamer in J. Chem. Theory Comput. where 3DNA was used extensively. This work represents a re-visit of the classic Dickerson−Drew B-DNA dodecamer d-[CGCGAATTCGCG]2 using state-of-the-art MD simulations with different ionic conditions and solvation models, and compares the MD trajectories with modern crystallographic and NMR data. Among the author list (Tomas Drsata, Alberto Perez, Modesto Orozco, Alexandre Morozov, Jiri Sponer, and Filip Lankas) are some well-known figures in the MD field of nucleic acid structures.
Reading through the text, I am not sure if the newly available functionality of x3dna_ensemble was used. From the excerpts of the citations given below, however, it seems obvious that 3DNA is now well-accepted by the MD community.
Snapshots taken in 10 ps intervals were analyzed using the 3DNA program.43 From 3DNA outputs, time series of conformational parameters were extracted. These included the intra-base-pair coordinates (buckle, propeller, opening, shear, stretch, and stagger), inter-base-pair or step coordinates (tilt, roll, twist, shift, slide, and rise) as well as groove widths (based on P−P distances), backbone torsions, and sugar puckers.
Contrary to the original work of Lankas et al.,31 the intra-base-pair and step coordinates used here are those defined by 3DNA.43
Here, we apply this model together with the 3DNA definitions of the intra-base-pair and step coordinates.43
However, important differences remain, and non- negligible differences are in fact observed between individual experimental structures also in the central part of DD, even though the intra-base-pair and step coordinates are computed using the same coordinate definitions64 (we consistently use the 3DNA coordinates in this work).

Glycosidic bond “is a type of covalent bond that joins a carbohydrate (sugar) molecule to another group, which may or may not be another carbohydrate.” In nucleic acid structures, the other group is a nucleobase, and the predominated type is the N-glycosidic bond where the purine (A/G) N9 or pyrimidine (C/T/U) N1 atom connects to the C1′ atom of the five-membered (deoxy) ribose sugar ring. Another well-known type is the C-glycosidic bond in pseudouridine, the most common modified base in RNA structures where the C5 atom instead of N1 is linked to the C1′ atom of the sugar ring.

Recently, I performed a survey of all nucleic-acid-containing structures in the PDB/NDB database to see how many types of glycosidic bond are there. As always, I noticed some inconsistencies in the data: nucleotides with disconnected base/sugar, a base labeled as U but with pseudoU-type C-glycosidic bond. Shown below are a few unusual types of glycosidic bond in otherwise seemingly “normal” structures:
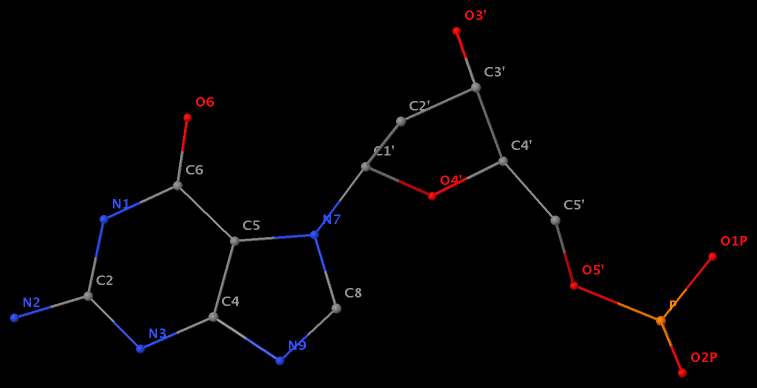
- The residue GN7 (number 28 on chain A) in PDB entry 1gn7 contains a N7-glycosylated guanine.
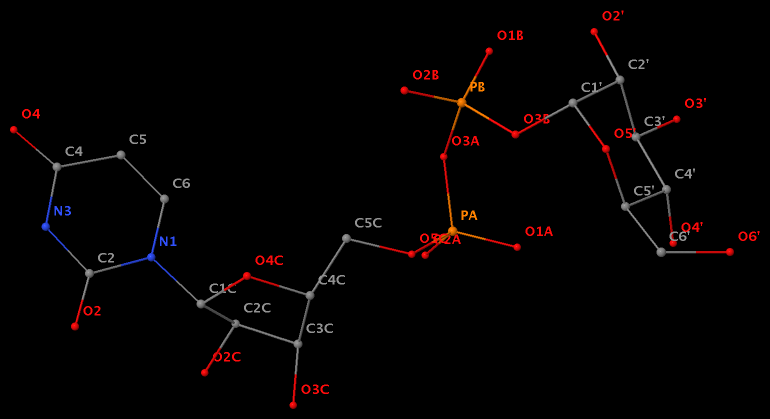
- The residue UPG (number 501 on chain A) in PDB entry 1y6f has sugar C1C (instead of C1′) atom connects to N1 of U.
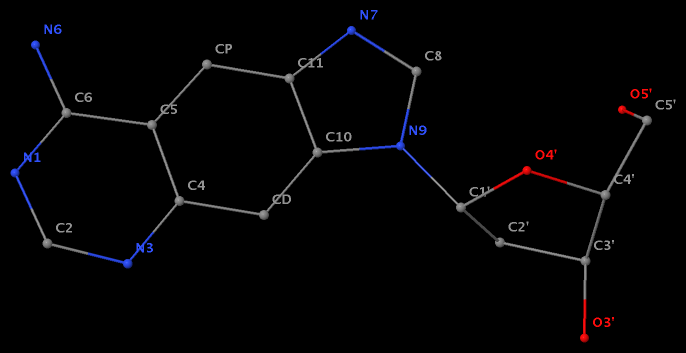
- The residue XAE (number 11 on chain B) in PDB entry 2icz contains a benzo-homologous adenine.
- The residue F5H (number 206 on chain B) in PDB entry 3v06 has N1 of U connects to C2′ of a six-membered sugar ring.
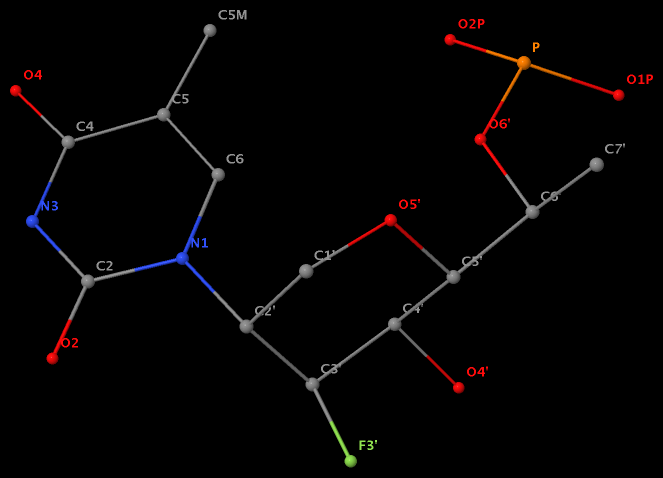
The unusual glycosidic bond has implications in 3DNA calculated parameters, for example the chi torsion angle. Identifying such cases would help refine 3DNA to provide sensible parameters and to avoid possible misinterpretations.













