Recently I came across the ligand thiamine pyrophosphate (TPP) in some RNA riboswitch structures. I was a bit surprised by the atom names adopted for the ligand by the PDB. See figures below for the chemical structure of TPP from the RCSB PDB website (first), and the three-dimensional structure of the ligand from the riboswitch 2gdi (second).

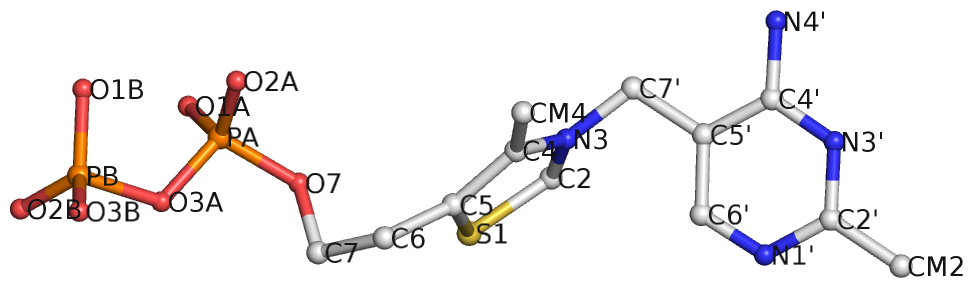
Specifically, the planar base-like moiety at the right has atom names ending with prime. To my knowledge, only sugar atom names of DNA and RNA nucleotides have the prime suffix, such as the 2′-hydroxyl group in RNA.
The RCSB webpage for TPP shows that currently there are 107 entries in the PDB, among which 100 are from proteins, 6 from RNA, and one in a RNA-protein complex. It is not clear to me whether the prime-bearing names in TPP are following any documented ‘standard’ or convention. DSSR is nevertheless taking a note of such ‘weird’ cases.

As of today, the number of registered users on the 3DNA Forum has reached 2000. Over the past three years, the annual average of resignations is 650, corresponding to approximately 1.8 per day. While many registrations use free email services (gmail, hotmail or yahoo, etc), a significant portion (especially more recent ones) employs their job email (e.g., .edu). This is clear sign of increasing trust the community puts in the Forum.
To ensure the 3DNA Forum spam-free, I’ve adhered a zero-tolerance policy of any trolling or suspicion activities. The anti-spam software has played a big role in making this clean status feasible, as is evident from the note: “120,933 Spammers blocked up until today”.
From a scientific perspective, all posted questions have been addressed promptly, normally within hours. Instead of feeling like a burden, maintaining the Forum and answering user questions have been a pleasure. I’d love to see more questions or posts on the Forum.

With great pleasure, I read the following annoancement from Rajendra Kumar on the 3DNA Forum:
Re: do_x3dna: a tool to analyze DNA/RNA in molecular dynamics trajectories
« Reply #1 on: Today at 10:53:31 AM »
Hello,
I have now made a new website for do_x3dna
(http://rjdkmr.github.io/do_x3dna). This website contains detailed
documentation for do_x3dna program and Python APIs.
Documentation for Python API is now available
(http://rjdkmr.github.io/do_x3dna/apidoc.html).
Few tutorials about the Python APIs are also now available
(http://rjdkmr.github.io/do_x3dna/tutorial.html).
Thanks.
With best regards,
Rajendra
Browsing through the do_x3dna website, I am impressed by the extensive documentation and tutorial. Clearly, do_x3dna has pushed the boundaries (in applicability and documentation) of the x3dna_ensemble Ruby script distributed with 3DNA v2.1.
As noted in GitHub page, do_x3dna has been developed to analyze fluctuations in DNA or RNA structures in molecular dynamics (MD) trajectories. It can be used for GROMACS MD trajectories, as well as those from NAMD and AMBER. It leaves no doubt that do_x3dna will boost 3DNA’s applications in the increasingly active field of DNA/RNA MD simulations.

From the very first release up until recently, the DSSR distribution had included two executables for Windows: one version was compiled on MinGW/MSYS, and the other on Cygwin. The executables are supposed to be run under the corresponding shells of the two environments respectively.
Since DSSR is a simple self-contained command-line tool, the MinGW/MSYS version also works directly under the Command Prompt of native Windows. So Windows users had the following three options to use DSSR:
- Download the MinGW/MSYS version to run it under the Command Prompt of native Windows. No need to install MinGW/MSYS.
- Download the MinGW/MSYS version to run it under the MinGW/MSYS environment, which must be installed separately.
- Download the Cygwin version to run it under the Cygwin environment, which must be installed separately.
Over times, I have observed some confusions among DSSR users as to which of the two executables to use on Windows. Luckily, I noticed by chance recently that the DSSR executable compiled under MinGW/MSYS runs just fine in the Cygwin shell. So as of v1.1.0-2014apr09, the DSSR distribution contains only one executable for Windows: compiled under MinGW/MSYS on 32-bit Windows XP, the same DSSR executable runs under the Command Prompt of native Windows, MinGW/MSYS, and Cygwin, either on a 32-bit or 64-bit Windows (XP, Vista, 7 or 8) machine.
A size fits all: I no longer need to provide two compiled versions of DSSR for Windows, and users have just one executable to download (no more space for confusions).
Note added on 2024-11-25: DSSR is distributed by the CTV (Columbia Technology Ventures). See https://home.x3dna.org

In addition to VARNA, the draw program in the RNAstructure package from the Mathews Laboratory can also be used to depict DSSR-derived RNA secondary structures in connect table (.ct) format. The draw program produces images in PostScript (or svg) format, in different styles from those generated by VARNA. Given below are a couple of examples on how to connect DSSR with draw.
The secondary structure of the PDB entry 1msy in DSSR-derived .ct file is as below:
27 DSSR-derived secondary structure in '1msy'
1 U 0 2 0 2647
2 G 1 3 26 2648
3 C 2 4 25 2649
4 U 3 5 24 2650
5 C 4 6 23 2651
6 C 5 7 22 2652
7 U 6 8 0 2653
8 A 7 9 0 2654
9 G 8 10 0 2655
10 U 9 11 0 2656
11 A 10 12 0 2657
12 C 11 13 17 2658
13 G 12 14 0 2659
14 U 13 15 0 2660
15 A 14 16 0 2661
16 A 15 17 0 2662
17 G 16 18 12 2663
18 G 17 19 0 2664
19 A 18 20 0 2665
20 C 19 21 0 2666
21 C 20 22 0 2667
22 G 21 23 6 2668
23 G 22 24 5 2669
24 A 23 25 4 2670
25 G 24 26 3 2671
26 U 25 27 2 2672
27 G 26 0 0 2673
Let the DSSR-derived .ct file for 1msy be named 1msy.ct, the following two draw-command runs will produce the secondary structure in PostScript (1msy.eps) and svg (1msy.svg) respectively.
draw 1msy.ct 1msy.eps
draw 1msy.ct 1msy.svg --svg -n 1
![1msy [GUAA tetra loop] 2nd structure produced with the RNAstructure 'draw' program 1msy [GUAA tetra loop] 2nd structure produced with the RNAstructure 'draw' program](http://forum.x3dna.org/images/1msy.svg)
The PDB entry 1ehz (yeast phenylalanine tRNA) has a pseudo knot, so the draw program will create a ‘circularized’ structure as shown below:
![1ehz [yeast phenylalanine tRNA] 2nd structure produced with the RNAstructure 'draw' program 1ehz [yeast phenylalanine tRNA] 2nd structure produced with the RNAstructure 'draw' program](http://forum.x3dna.org/images/1ehz.svg)
Note the following two caveats:

Recently I was surprised by some cases of nucleotides with missing atoms in PDB entry 1pns. The story started like this: 3DNA/DSSR maps various nucleotide names to one-letter codes, based on the data file baselist.dat (see post Modified nucleotides in the PDB). In the meantime, 3DNA/DSSR internally assigns a nucleotide as either purine or pyrimidine, by virtue of coordinates of base atoms. Be definition, purines should only include A/a/G/g/I/i, and pyrimidines C/c/T/t/U/u/P/p. However, no consistency check has been implemented in DSSR until just now.
I first noticed the inconsistency between residue name and atom coordinates for nucleotide A6 on chain U (hereafter referred to as U.A6) in 1pns. The nucleotide has standard name ‘ A’, obviously a purine. However, somehow DSSR classified it as a pyrimidine based on atomic coordinates. Upon further check of the PDB data file, I found the following remarks:
REMARK 470 MISSING ATOM
REMARK 470 THE FOLLOWING RESIDUES HAVE MISSING ATOMS(M=MODEL NUMBER;
REMARK 470 RES=RESIDUE NAME; C=CHAIN IDENTIFIER; SSEQ=SEQUENCE NUMBER;
REMARK 470 I=INSERTION CODE):
REMARK 470 M RES CSSEQI ATOMS
REMARK 470 A U 6 N9 C8 N7
REMARK 470 G U 8 N9 C8 N7
REMARK 470 A U 12 N9 C8 N7
REMARK 470 A U 13 N9 C8 N7
REMARK 470 A U 14 N9 C8 N7
The atomic coordinates for U.A6 are as below:
ATOM 34447 P A U 6 81.861 37.210 78.651 1.00378.87 P
ATOM 34448 OP1 A U 6 80.631 37.121 77.831 1.00378.87 O
ATOM 34449 OP2 A U 6 81.665 37.221 80.119 1.00378.87 O
ATOM 34450 O5' A U 6 82.707 38.495 78.212 1.00378.87 O
ATOM 34451 C5' A U 6 83.948 38.777 78.887 1.00378.87 C
ATOM 34452 C4' A U 6 84.600 40.000 78.276 1.00378.87 C
ATOM 34453 O4' A U 6 84.975 39.698 76.901 1.00378.87 O
ATOM 34454 C3' A U 6 83.714 41.239 78.153 1.00378.87 C
ATOM 34455 O3' A U 6 83.654 41.968 79.369 1.00378.87 O
ATOM 34456 C2' A U 6 84.403 42.015 77.020 1.00378.87 C
ATOM 34457 O2' A U 6 85.564 42.655 77.474 1.00378.87 O
ATOM 34458 C1' A U 6 84.834 40.864 76.105 1.00378.87 C
ATOM 34459 C5 A U 6 82.033 39.296 74.209 1.00378.87 C
ATOM 34460 C6 A U 6 82.941 39.553 75.166 1.00378.87 C
ATOM 34461 N6 A U 6 81.170 39.949 72.090 1.00378.87 N
ATOM 34462 N1 A U 6 83.830 40.588 75.041 1.00378.87 N
ATOM 34463 C2 A U 6 83.843 41.410 73.939 1.00378.87 C
ATOM 34464 N3 A U 6 82.899 41.124 72.974 1.00378.87 N
ATOM 34465 C4 A U 6 81.968 40.108 73.016 1.00378.87 C
No atom records for N7, C8 and N9. So far, so good. However, surprise came when I visualized U.A6 in Jmol, as shown in the following image. Note here atom N1 is connected to C1’ as in pyrimidines, and N6 is bonded to C4!
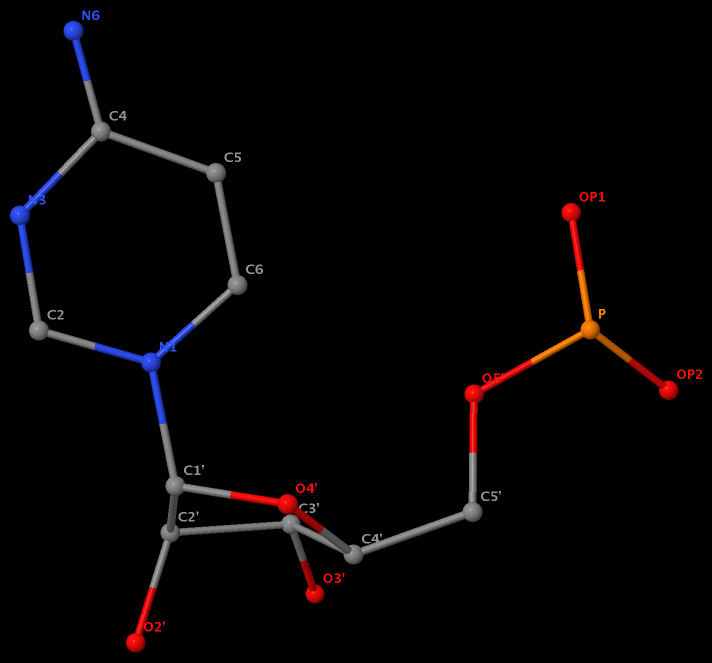
The same issue also exists for U.G8 (see figure below), U.A12, U.A13, and U.A14.
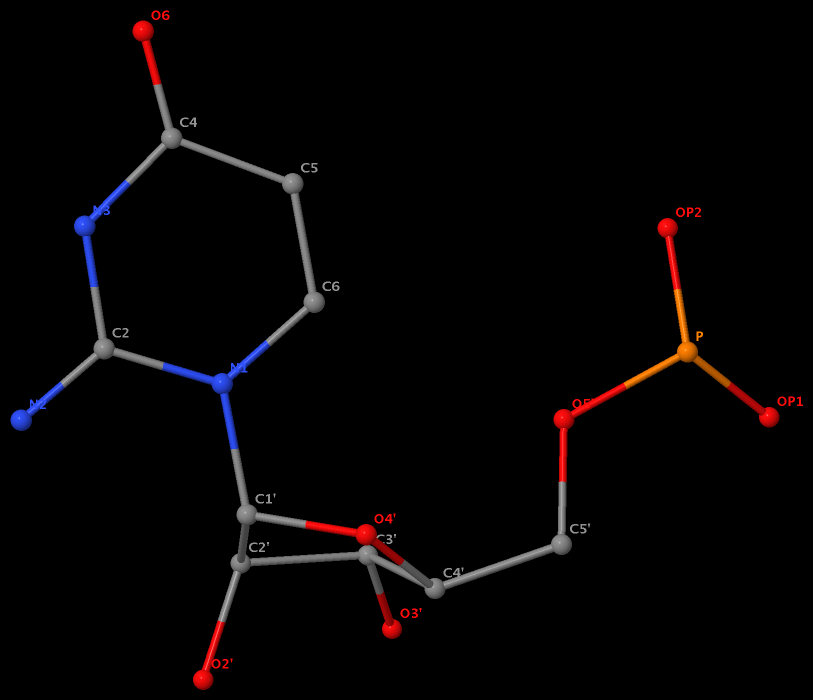
It is beyond my imagination to understand why such weird cases exist in the PDB, even given the lousy resolution (8.7 Å) of 1pns.

I recently upgraded my Macs to OS X Mavericks to check if 3DNA/DSSR works in the new operating system. I am glad to report that both run without a hitch, as expected.
Since OS X Mavericks is free from the Mac App Store, it will quickly become the de facto version virtually all Mac users would use. I also noticed that Ruby on Mavericks has been upgraded to ruby 2.0.0p247 (2013-06-27 revision 41674), a major step forward from the now retiring Ruby 1.8.7 distributed in previous versions of Mac OS X.
As a rule, I’d ensure that 3DNA/DSSR executes properly in major releases of the commonly used operating systems — Mac, Windows, and Linux.

While having not used DOS for ages, I am glad to find that the DSSR version compiled for MinGW/MSYS on Windows works perfectly under this operating system (see screenshot below). The DSSR DOS command-line interface functions exactly the same as for Linux, Mac OS X, MinGW/MSYS, and CygWin. Among other possible usages, it allows for batch files to take advantage of DSSR.
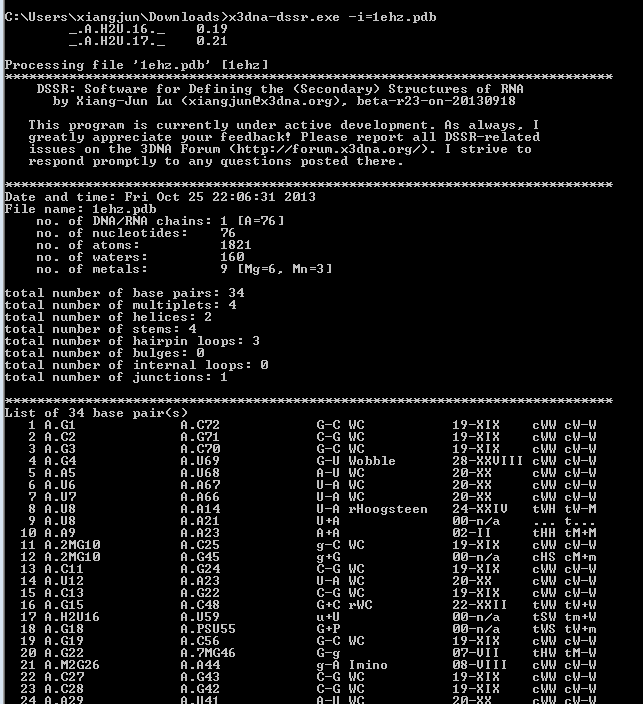
Implementing DSSR in strict ANSI C as a self-contained and zero-dependent command-line program pays off enormously: it simplifies code maintenance and ensures that the program is applicable wherever a C compiler exists. The easy web interface to DSSR makes the program universally accessible.

Aside from its extensive functionality for RNA structural analyses, DSSR also introduces a consistent and flexible way to process command-line options. Here, each option can be specified via a --key[=value] pair (or -key[=value] or key[=value]; i.e., two/one/zero preceding dashes are all accepted), key can be in either lower, UPPER or MiXed case, and value is optional for Boolean switches. Furthermore, options can be put in any order; if the same key is repeated more than once, the value specified last overwrites corresponding previous settings.
As always, the rules are best illustrated with concrete examples. Some typical use-cases are given below:
#1 analyze PDB entry '1msy', with default output to stdout
x3dna-dssr --input=1msy.pdb
#2 same as #1, with output directed to file '1msy.out'
x3dna-dssr --input=1msy.pdb --output=1msy.out
#3-6, same as #2
x3dna-dssr --output=1msy.out --input=1msy.pdb
x3dna-dssr --OUTPUT=1msy.out --Input=1msy.pdb
x3dna-dssr -output=1msy.out input=1msy.pdb
x3dna-dssr output=1msy.out --input=1msy.pdb
#7 the value '1ehz.pdb' overwrites '1msy.pdb'
x3dna-dssr --input=1msy.pdb input=1ehz.pdb
#8-12 with the switch --more set to true
x3dna-dssr -input=1msy.pdb --more
x3dna-dssr -input=1msy.pdb --more=true
x3dna-dssr -input=1msy.pdb --more=yes
x3dna-dssr -input=1msy.pdb --more=on
x3dna-dssr -input=1msy.pdb --more=1
#13 same as without specifying --more,
# or with values set to false/no/0
x3dna-dssr -input=1msy.pdb --more=off
#14 shorthand forms for --input and --output
x3dna-dssr -i=1msy.pdb -o=1msy.out
#15 it can also be more verbose
x3dna-dssr --input-pdb-file=1msy.pdb
#16-18 within a key, separator dash(-) and underscore (_)
# are treated the same, and can be omitted
x3dna-dssr -i=1msy.pdb -non-pair
x3dna-dssr -i=1msy.pdb -non_pair
x3dna-dssr -i=1msy.pdb -nonpair
By allowing for 2/1/0 dashes to precede each key and a dash/underscore character or none to separate words within the key, DSSR provides users with great flexibility in specifying command-line options to fit into their preferred styles. Not surprisingly, new programs to be added into 3DNA, or the version 3 release of the software will all follow the same convention.




![1msy [GUAA tetra loop] 2nd structure produced with the RNAstructure 'draw' program 1msy [GUAA tetra loop] 2nd structure produced with the RNAstructure 'draw' program](http://forum.x3dna.org/images/1msy.svg)
![1ehz [yeast phenylalanine tRNA] 2nd structure produced with the RNAstructure 'draw' program 1ehz [yeast phenylalanine tRNA] 2nd structure produced with the RNAstructure 'draw' program](http://forum.x3dna.org/images/1ehz.svg)


