Crystal structure of SARS-CoV-2 stem–loop 5 (SL5) (PDB id: 9E9Q; Jones CP, Ferré-D'Amaré AR. 2025. Crystallographic and cryoEM analyses reveal SARS-CoV-2 SL5 is a mobile T-shaped four-way junction with deep pockets. RNA 31: 949–960). The T-shaped four-way junction of the coronavirus SL5 structural element provides a starting point for examining the structures of larger RNA motifs and their interactions with other molecules. Image highlighting the four arms of the junction. The RNA backbone is depicted by a gray ribbon. The bases within the arms of the junction are colored respectively in blue, red, yellow, and cyan. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for Structural Bioinformatics of Nucleic Acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
As the developer of DSSR, I am thrilled to see its application in cutting-edge research across multiple disciplines. Below is a list of four recent publications that highlight how DSSR has been utilized, underscoring its versatility and significance in structural bioinformatics.
In the Geng et al. (2025) Nucleic Acids Research (NAR) paper, titled 'Revealing hidden protonated conformational states in RNA dynamic ensembles', DSSR is simply cited as follows:
All bp geometries, hydrogen-bond, backbone, stacking, and sugar dihedral angles were calculated using X3DNA-DSSR [77].
In the preprint by Gordan et al. (2025), titled 'High-throughput characterization of transcription factors that modulate UV damage formation and repair at single-nucleotide resolution', DSSR is cited as follows:
Step base stacking, base pair shift, base pair slide, interbase angle, pseudorotation angle, and sugar puckering classifications of nucleobases were computed using X3DNA-DSSR (v2.5.0)75. Base stacking was defined as the overlapping polygon area in Å2 when projecting the dipyrimidine base ring atoms (excluding exocyclic atoms) into the mean base pair plane76. The sugar ring pseudorotation phase angle of each pyrimidine was also calculated using X3DNA-DSSR as described by Altona, C. & Sundaralingam, M.77 Interbase angle was defined as sqrt(propeller2+buckle2) per the X3DNA-DSSR documentation.
Figure 6: TF Binding Induces Structural Distortion Favorable to UV Dimerization is highly informative, particularly panel (a), which illustrates the ensemble of structural parameters that predispose dipyrimidines to cyclobutane pyrimidine dimers (CPD) or 6-4 pyrimidine-pyrimidones (6-4 PP) formation. DSSR is designed as an integrated software tool, offering a comprehensive suite of structural parameters not found in any other single tool I am aware of. Despite this, the innovative use of DSSR by Gordan et al. exceeds my expectations and demonstrates its versatility.
In the preprint by Kubaney et al. (2025) from the Baker group, titled 'RNA sequence design and protein-DNA specificity prediction with NA-MPNN', DSSR is cited as follows:
On the pseudoknot subset, we evaluate additional structure‐ and reactivity‐based metrics. DSSR v2.3.241 is used to extract the ground‐truth secondary structure from the native crystal structures. For each designed sequence, RibonanzaNet predicts 2A3 reactivity profiles, from which we compute predicted OpenKnot scores (see https://github.com/eternagame/OpenKnotScore)31 using the predicted reactivity together with the DSSR ground truth.
In a recent NSMB paper from the Baker group, titled 'Computational design of sequence-specific DNA-binding proteins', 3DNA is cited as follows:
RIF docking of scaffolds onto DNA targets (DBP design step 1) Structures of B-DNA for each target (Supplementary Table 2) were generated by (1) using the DNA portion of PDB 1BC8 (ref. 60), PDB 1YO5 (ref. 61), PDB 1L3L (ref. 51) or PDB 2O4A (ref. 62) or (2) using the software X3DNA63, followed by a constrained Rosetta relax of the DNA structure.
Please note that 3DNA has been replaced by DSSR. The functionality for constructing B-DNA models, previously provided by 3DNA, is now directly available in DSSR via its fiber and rebuild modules.
In the preprint by Si et al. (2025), titled 'End-to-End Single-Stranded DNA Sequence Design with All-Atom Structure Reconstruction', DSSR is cited as follows:
Since ViennaRNA and NUPACK require secondary structures as input, we used DSSR35 to extract secondary structures from the corresponding ssDNA three-dimensional structures.
The above use cases are merely a sample of how DSSR is utilized in the scientific literature. It is reasonable to state that DSSR has emerged as a de facto standard tool within the field of nucleic acid structural bioinformatics. Overall, DSSR is a mature, robust, and efficient software product that is actively developed and maintained. I am committed to making DSSR synonymous with quality and value. Its unmatched functionality, usability, and support save users significant time and effort compared to alternative solutions.
DSSR is available free of charge for academic users. Additionally, it has been integrated into other high-profile bioinformatics resources, including NAKB, PDB-redo, and N•ESPript.
References
- Geng A, Roy R, Ganser L, Li L, Al-Hashimi HM. Revealing hidden protonated conformational states in RNA dynamic ensembles. Nucleic Acids Research. 2025;53:gkaf1366. https://doi.org/10.1093/nar/gkaf1366.
- Gordan R, Wasserman H, Chi B, Bohm K, Duan M, Sahay H, et al. High-throughput characterization of transcription factors that modulate UV damage formation and repair at single-nucleotide resolution. 2025. https://doi.org/10.21203/rs.3.rs-8197218/v1.
- Kubaney A, Favor A, McHugh L, Mitra R, Pecoraro R, Dauparas J, et al. RNA sequence design and protein–DNA specificity prediction with NA-MPNN. 2025. https://doi.org/10.1101/2025.10.03.679414.
- Glasscock CJ, Pecoraro RJ, McHugh R, Doyle LA, Chen W, Boivin O, et al. Computational design of sequence-specific DNA-binding proteins. Nat Struct Mol Biol. 2025;32:2252–61. https://doi.org/10.1038/s41594-025-01669-4.
- Si Y, Xu Y, Chen L. End-to-end single-stranded DNA sequence design with all-atom structure reconstruction. 2025. https://doi.org/10.64898/2025.12.05.692525.
DSSR 2.0 is out. It integrates an unprecedented set of features into one computational tool, including analysis/annotation, schematic visualization, and model building of 3D nucleic acid structures. DSSR 2.0 supersedes 3DNA 2.4, which is still maintained but no additional features other than bug fixes are scheduled. See the DSSR 2.0 overview PDF.
DSSR delivers a great user experience by solving problems and saving time. Considering its usability, interoperability, features, and support, DSSR easily stands out among `competitors’. It exemplifies a `solid software product’. I strive to make DSSR a pragmatic tool that the structural bioinformatics community can count on.
DSSR 2.0 is licensed by Columbia University. The software remains free for academic users, with the basic user manual. The professional user manual (over 230 pages, including 7 appendices) is available for paid academic users or commercial users only. Licensing revenue helps ensure the long-term sustainability of the DSSR project.
Additionally, the paper “DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL” has recently been published in Nucleic Acids Research, 48(13):e74. Check the web interface.
The DSSR-PyMOL paper/website has been rated “very good” and classified as “Good for Teaching”. See Vicens Q: Faculty Opinions Recommendation of [Lu XJ, Nucleic Acids Res 2020 48(13):e74]. In Faculty Opinions, 14 Aug 2020; 10.3410/f.738001682.793577327

Recently I performed a survey of citations to thirteen 3DNA-related publications using Web of Science from Clarivate Analytics. The time range is from 2015 to 2020 (June 30), for a total of five-and-half years. The 1,050 citations span 223 scientific journals, covering a broad range of research fields such as biology, medicine, chemistry, physics, materials etc. Not surprisingly, the citing journals include Cell, Nature and sub-journals, Science, and PNAS.
Each of following six papers has been cited over 50 times, as detailed below. Adding the six numbers together, there are 962 citations, accounting for 92% of the total 1,050.
- [
402 times in 138 journals] Lu,X.-J. and Olson,W.K. (2003) 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res., 31, 5108–5121.
- [
201 times in 81 journals] Lu,X.-J. and Olson,W.K. (2008) 3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc., 3, 1213–1227.
- [
127 times in 71 journals] Zheng,G., Lu,X.J. and Olson,W.K. (2009) Web 3DNA––a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res, 37, W240-6.
- [
115 times in 57 journals] Olson,W.K., Bansal,M., Burley,S.K., Dickerson,R.E., Gerstein,M., Harvey,S.C., Heinemann,U., Lu,X.-J., Neidle,S., Shakked,Z., Sklenar,H., Suzuki,M., Tung,C.-S., Westhof,E., Wolberger,C. and Berman,H.M. (2001) A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol., 313, 229–237.
- [
66 times in 32 journals] Lu,X.-J., Bussemaker,H.J. and Olson,W.K. (2015) DSSR: An integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res., 43, e142.
- [
51 times in 41 journals] Lu,X.J., Shakked,Z. and Olson,W.K. (2000) A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol., 300, 819–40.
The top 21 journals that cite 3DNA papers 10 times or more are listed below. Nucleic Acids Research stands out, with a total of 148 citations, accounting for 14% of the total 1,050 citations.
148 Nucleic Acids Research
84 Journal of Physical Chemistry B
40 Physical Chemistry Chemical Physics
34 Biophysical Journal
33 Journal of Chemical Theory and Computation
29 Biochemistry
29 RNA
24 PLoS One
24 Scientific Reports
22 Journal of Biomolecular Structure & Dynamics
20 Bioinformatics
19 Journal of Chemical Information and Modeling
16 Nature Communications
15 Biopolymers
15 Journal of the American Chemical Society
12 Acta Crystallographica Section D: Structural Biology
12 Journal of Molecular Modeling
11 Chemistry: a European Journal
11 Journal of Chemical Physics
10 Journal of Biological Chemistry
10 Structure

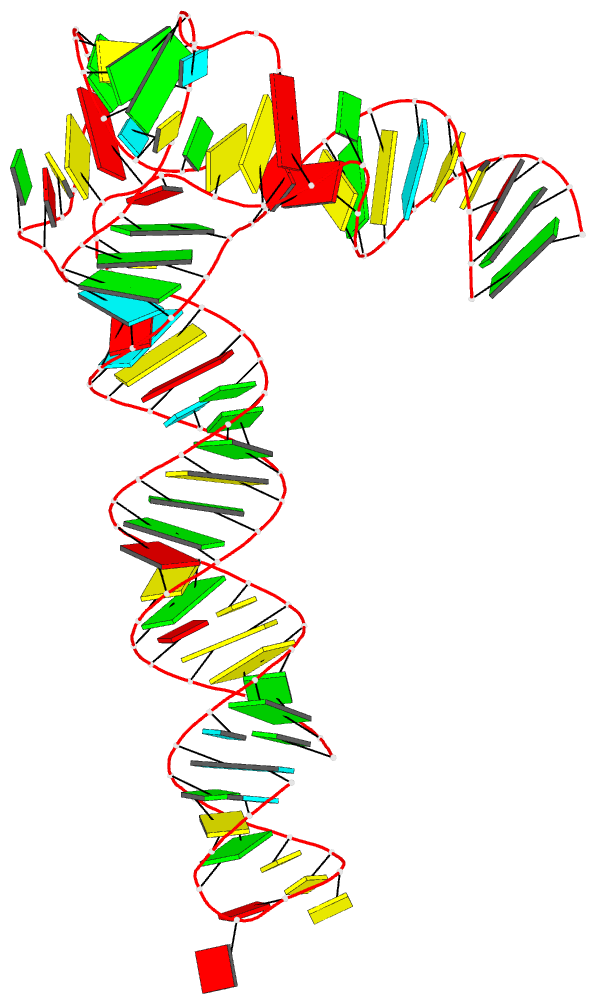
Following my previous post 3DNA/blocview-PyMOL images in covers of the RNA journal in 2019, here is an update for 2020. The cover images of the January to July issues have all been generated with help of 3DNA and provided by the NDB:
RNA is displayed as a red ribbon; block bases use NDB colors: A—red, C—yellow, G—green, U—cyan. The image was generated using 3DNA/blocview and PyMol software. Cover image provided by the Nucleic Acid Database (ndbserver.rutgers.edu).
Here is the composite figure of the seven cover images, with the brand new DSSR-PyMOL schematics for comparison.
Details of the seven structures illustrated in the cover images are described below:
- January 2020 Pumilio homolog PUF domain in complex with RNA (PDB id: 5yki; Zhao YY, Mao MW, Zhang WJ, Wang J, Li HT, Yang Y, Wang Z, Wu JW. 2018. Expanding RNA binding specificity and affinity of engineered PUF domains. Nucleic Acids Res 46: 4771–4782). Engineered nine-repeat PUF domain binds to its RNA target specifically and with high binding affinity.
- February 2020 Aprataxin RNA–DNA deadenylase product complex (PDB id: 6cvo; Tumbale P, Schellenberg MJ, Mueller GA, Fairweather E, Watson M, Little JN, Krahn J, Waddell I, London RE, Williams RS. 2018. Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease. EMBO J 37: e98875). Human aprataxin RNA–DNA deadenylase protects genome integrity and corrects abortive DNA ligation arising during ribonucleotide excision repair and base excision DNA repair.
- March 2020 PreQ1 riboswitch (PDB id: 6e1w; Connelly CM, Numata T, Boer RE, Moon MH, Sinniah RS, Barchi JJ, Ferre-D’Amare AR, Schneekloth Jr JS. 2019. Synthetic ligands for PreQ1 riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure. Nat Commun 10: 1501). Class I PreQ1 riboswitch regulates downstream gene expression in response to its cognate ligand PreQ1 (7-aminomethyl-7-deazaguanine).
- April 2020 Hatchet ribozyme (PDB id: 6jq6; Zheng L, Falschlunger C, Huang K, Mairhofer E, Yuan S, Wang J, Patel DJ, Micura R, Ren A. 2019. Hatchet ribozyme structure and implications for cleavage mechanism. Proc Natl Acad Sci 116: 10783–10791). This crystal structure of the hatchet ribozyme product features a compact symmetric dimer.
- May 2020 Adenovirus virus-associated RNA (PDB id: 6ol3; Hood IV, Gordon JM, Bou-Nader C, Henderson FE, Bahmanjah S, Zhang J. 2019. Crystal structure of an adenovirus virus-associated RNA. Nat Commun 10: 2871). Acutely bent viral RNA fragment is a protein kinase R inhibitor and features an unusually structured apical loop, a wobble-enriched, coaxially stacked apical and tetra-stems, and a central domain pseudoknot that resembles codon-anticodon interactions.
- June 2020 Archeoglobus fulgidus L7Ae bound to cognate K-turn (PDB id: 6hct; Huang L, Ashraf S, Lilley DMJ. 2019. The role of RNA structure in translational regulation by L7Ae protein in archaea. RNA 25: 60–69). 50S archaeal ribosome protein L7Ae binds to a K-turn structure in the 5′-leader of the mRNA of its structural gene to regulate translation.
- July 2020 Spinach RNA aptamer/Fab complex (PDB id: 6b14; Koirala D, Shelke SA, Dupont M, Ruiz S, DasGupta S, Bailey LJ, Benner SA, Piccirilli JA. 2018. Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography. Nucleic Acids Res 46: 2624–2635). Novel Fab-RNA module can serve as an affinity tag for RNA purification and imaging and as a chaperone for RNA crystallography.

The paper, titled DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL, has just been published in Nucleic Acids Research (online on May 22, 2020). Here is the abstract:
Sophisticated analysis and simplified visualization are crucial for understanding complicated structures of biomacromolecules. DSSR (Dissecting the Spatial Structure of RNA) is an integrated computational tool that has streamlined the analysis and annotation of 3D nucleic acid structures. The program creates schematic block representations in diverse styles that can be seamlessly integrated into PyMOL and complement its other popular visualization options. In addition to portraying individual base blocks, DSSR can draw Watson-Crick pairs as long blocks and highlight the minor-groove edges. Notably, DSSR can dramatically simplify the depiction of G-quadruplexes by automatically detecting G-tetrads and treating them as large square blocks. The DSSR-enabled innovative schematics with PyMOL are aesthetically pleasing and highly informative: the base identity, pairing geometry, stacking interactions, double-helical stems, and G-quadruplexes are immediately obvious. These features can be accessed via four interfaces: the command-line interface, the DSSR plugin for PyMOL, the web application, and the web application programming interface. The supplemental PDF serves as a practical guide, with complete and reproducible examples. Thus, even beginners or occasional users can get started quickly, especially via the web application at http://skmatic.x3dna.org.
A brief history on DNA/RNA schematics as implemented in SCHNAaP/SCHNArP, 3DNA, and now in DSSR:
The idea of representing bases and WC-pairs as rectangular blocks came from the pioneering work of Calladine et al. (27,28) The block schematics were first implemented in the pair of SCHNAaP/SCHNArP programs (29,30) for rigorous analysis and reversible rebuilding of double-helical nucleic acid structures. The algorithms that underpinned SCHNAaP/SCHNArP laid the foundation of ‘analyze’ and ‘rebuild’, two core components of the 3DNA suite of programs (31–33). 3DNA also takes advantage of the standard base reference frame (34), and comprises quite a few other related programs. One of them is ‘blocview’, a script which calls several 3DNA utility programs to generate individual base blocks and set the view, MolScript (35) to produce backbone ribbons, and Raster3D (36) to render the composite image. The 3DNA ‘blocview’ schematics catch characteristic attributes of nucleic acid structures. They have gradually become popular and been adopted into the RCSB PDB (1) and the NDB (37), and then propagated into other bioinformatics resources (e.g., the ‘RNA Structure Atlas’ website hosted by the Leontis-Zirbel RNA group).
DSSR supersedes ‘blocview’ by eliminating all the internal and external dependencies of the 3DNA utility program. DSSR produces block representations, not only of individual bases but also WC-pairs and G-tetrads, that can be fed directly into PyMOL. The DSSR-PyMOL integration is easier to use, has more features, and produces better schematics than the original 3DNA-blocview approach.
Indeed, the base block schematics have continuously evolved for over two decades, as appreciated in the acknowledgements:
I would like to thank Christopher A. Hunter, Christopher R. Calladine, Helen M. Berman, Catherine L. Lawson, Zukang Feng, Wilma K. Olson and Harmen J. Bussemaker for their helpful input on the block schematic during its continuous evolution for over two decades. I appreciate Thomas Holder (PyMOL Principal Developer, Schrödinger, Inc.) for writing the DSSR plugin for PyMOL, and for providing insightful comments on the manuscript and the web application interface. I also thank Jessalyn Lu and Yin Yin Lu for proofreading the manuscript, and the user community for feedback.
Notably, the supplemental PDF has been diligently written to serve as a practical guide, with complete and reproducible examples. In fact, the paper concludes with the following two sentences:
Finally, all results reported here are completely reproduceable (see the supplemental PDF). Any questions related to this work are welcome and will be openly addressed on the 3DNA Forum (http://forum.x3dna.org).

As of version 2.0 (to be released soon), DSSR has a new module for in silico base mutations that is context sensitive. Powered by the DSSR analysis engine, the module allows users to perform base mutations in unprecedented flexibility and convenance. Here are some examples:
- Mutate all bases in hairpin loops to a specific base (e.g., G)
- Mutate all non-stem bases to a specific base (e.g., U)
- Mutate bases 2-12 to a specific base (e.g., A) regardless of context
- Mutate bases 1-10 in a given structure to a new sequence (e.g., AUAUAUAUAU)
- Mutate all bases of the same type to another (e.g., A to G)
- Mutate all bases of the same type to another (e.g., C to U) except for some nucleotides
- Mutate all G-C Watson-Crick (WC) pairs to C-G WC pairs, and A-U to U-A
- Mutate all G-tetrads in G-quadruplexes to non-G-tetrads (e.g., U-tetrads)
By default, the mutation preserves both the geometry of the sugar-phosphate backbone and the base reference frame (position and orientation). As a result, re-analyzing the mutated model gives the same base-pair and step parameters as those of the original structure.
Over the years, the 3DNA mutate bases program has been cited in the literature and patent, including the following ones:
- Howe, John A., et al. Selective small-molecule inhibition of an RNA structural element. Nature 526.7575 (2015): 672-677.
- Wang, Hao, et al. Dual-targeting small-molecule inhibitors of the Staphylococcus aureus FMN riboswitch disrupt riboflavin homeostasis in an infectious setting. Cell Chemical Biology 24.5 (2017): 576-588.
- AlQuraishi, Mohammed, and Harley H. McAdams. Three enhancements to the inference of statistical protein‐DNA potentials. Proteins: Structure, Function, and Bioinformatics 81.3 (2013): 426-442.
- Wang, Harris, Sagi Shapira, and Victoria Stockman. High-throughput strategy for dissecting mammalian genetic interactions. U.S. Patent Application No. 15/747,677.
The DSSR mutation module has completely obsoleted the mutate_bases program distributed in 3DNA v2.x. In addition to serving as a drop-in replacement of mutate_bases, the DSSR approach offers much more features and versatility: it is simply better.

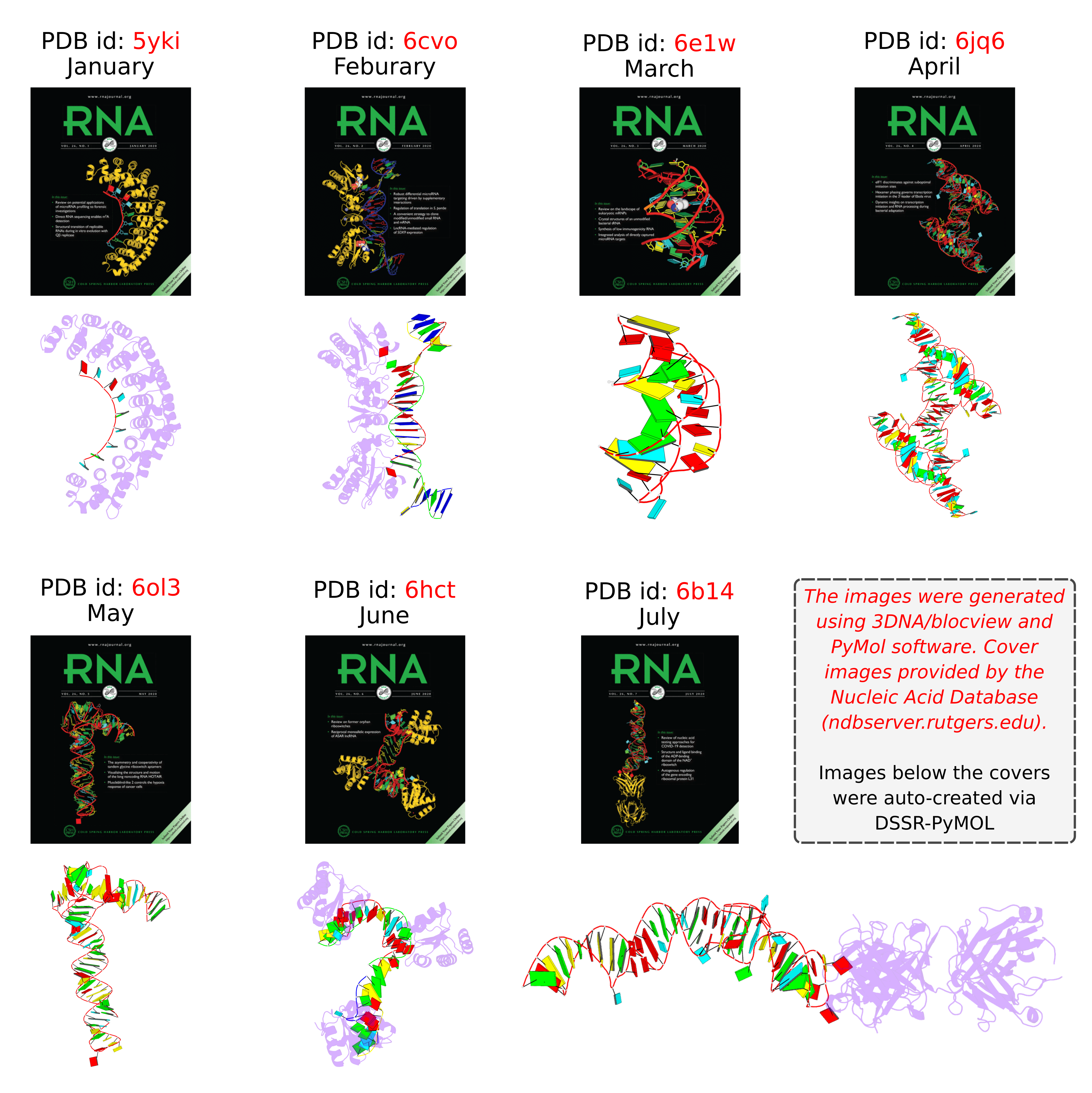
I recently noticed a bioRxiv preprint, titled Role of RNA Guanine Quadruplexes in Favoring the Dimerization of SARS Unique Domain in Coronaviruses by a European team consisting of scientists from France, Italy, and Spain. The abstract is as follows. Figure 1 shows a schematic representation of the mRNA with a G-Quadruplex structure, functioning in a healthy cell and an infected cell by coronavirus.
Coronaviruses may produce severe acute respiratory syndrome (SARS). As a matter of fact, a new SARS-type virus, SARS-CoV-2, is responsible of a global pandemic in 2020 with unprecedented sanitary and economic consequences for most countries. In the present contribution we study, by all-atom equilibrium and enhanced sampling molecular dynamics simulations, the interaction between the SARS Unique Domain and RNA guanine quadruplexes, a process involved in eluding the defensive response of the host thus favoring viral infection of human cells. The results obtained evidence two stable binding modes with guanine quadruplexes, driven either by electrostatic (dimeric mode) or by dispersion (monomeric mode) interactions, are proposed being the dimeric mode the preferred one, according to the analysis of the corresponding free energy surfaces. The effect of these binding modes in stabilizing the protein dimer was also assessed, being related to its biological role in assisting SARS viruses to bypass the host protective response. This work also constitutes a first step of the possible rational design of efficient therapeutic agents aiming at perturbing the interaction between SARS Unique Domain and guanine quadruplexes, hence enhancing the host defenses against the virus.
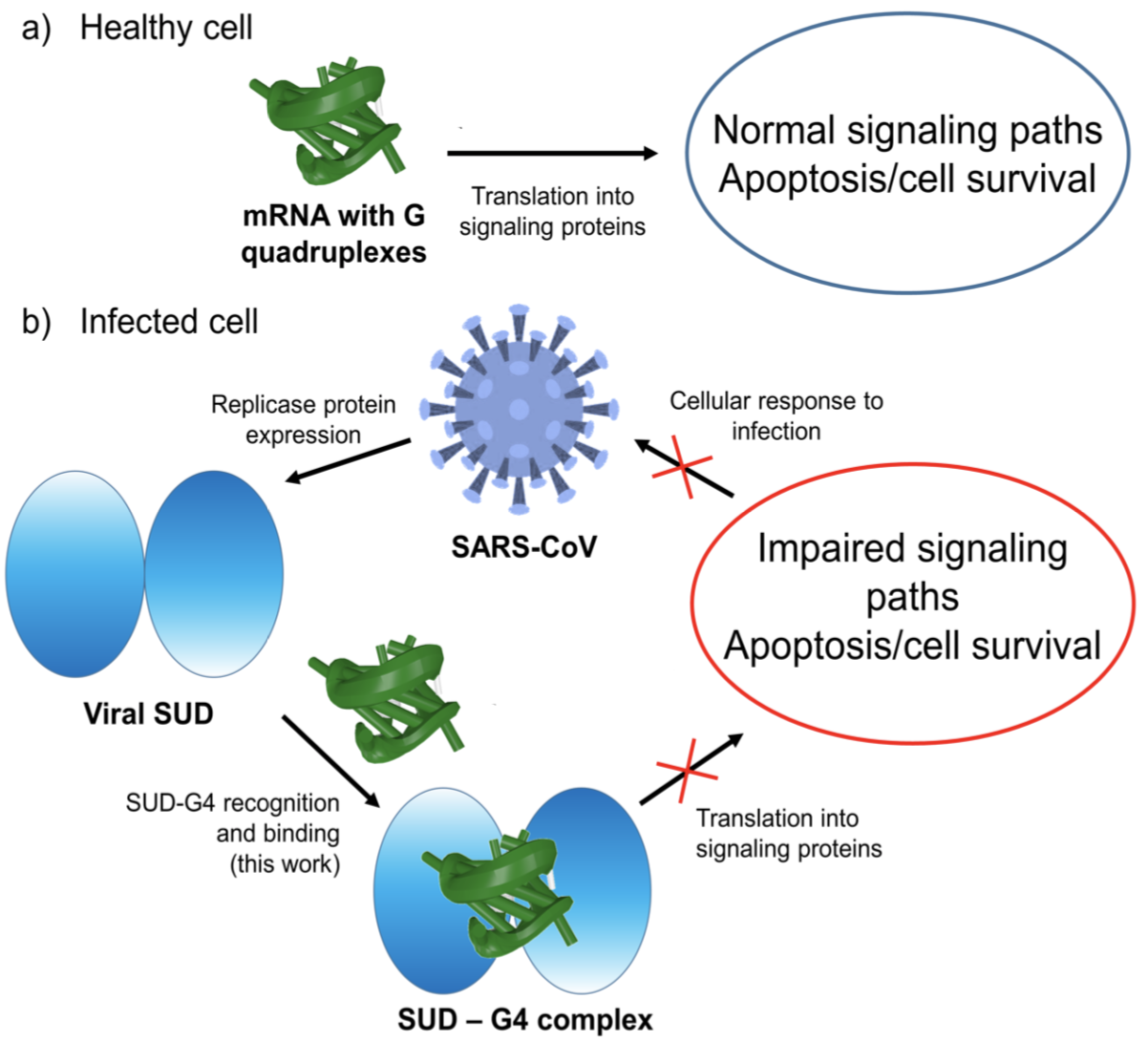
Figure 1) Schematic representation of the mRNA function in a) a healthy cell and b) an infected cell by coronavirus. Panel b) showcases the influence of viral SUD binding to G4 sequences of mRNA that encodes crucial proteins for the apoptosis/cell survival regulation and other signaling paths.
In the manuscript, the software tools employed in this MD study are described as below:
… Both protein and RNA have been described with the amber force field including the bsc1 corrections, and the MD simulations have been performed in the constant pressure and temperature ensemble (NPT) at 300K and 1 atm. All MD simulations have been performed using the NAMD code and analyzed via VMD, the G4 structure has also been analyzed with the 3DNA suite.
I am glad that 3DNA has played a role in the analysis of G-quadruplexes in this timely contribution. In particular, I would like to draw attention of the community to 3DNA-DSSR which has a brand-new module dedicated to the automatic identification and comprehensive characterization of G-quadruplexes. The DSSR-annotated G-quadruplexes from the PDB should be of great interest to a wide audience, especially the experimentalists. As a concrete example, the authors noted that “The crystal structure … of the oligonucleotide (pdb 1J8G) have been chosen coherently with the experimental work performed by Tan et al”. Follow the link to see results of DSSR-derived G-quadruplex features in PDB entry 1J8G and you are guaranteed to see features not available elsewhere.
Note added on July 9, 2020: This paper has been published in J. Phys. Chem. Lett. 2020, 11, 5661−5667.

The Kribelbauer et al. article, Towards a mechanistic understanding of DNA methylation readout by transcription factors has recently been published in the Journal of Molecular Biology (JMB). I am honored to be among the author list, and I learned a lot during the process. For the project, I added the --methyl-C (short-form: --5mc) option to SNAP (v1.0.6-2019sep30) for the automatic identification and annotation of DNA-transcription factor (TF) complexes containing 5-methyl-cytosine (5mC). The results are presented in a dynamic table, easily accessible at URL http://snap-5mc.x3dna.org, and summarized in Fig. 1 “Structural basis of how TFs recognize methylated DNA” (see below) of the JMB paper.

Details on the SNAP-enabled curation of TF-DNA complexes containing 5mC from atomic coordinates in the Protein Data Bank (PDB) are available in a tutorial page at http://snap-5mc.x3dna.org/tutorial. In essence, the process can be easily understood via a concrete example with PDB id 4m9e, as shown below.
x3dna-snap --methyl-C --type=base -i=4m9e.pdb -o=4m9e-5mC.out
Here the --methyl-C option is specific for 5mC-DNA, and --type=base ensures that at least one DNA base atom is contacting protein amino acid(s). If these conditions are fulfilled, SNAP would produce two additional 5mC-related files, apart from the normal output file (i.e., 4m9e-5mC.out, as specified in the example):
- 4m9e-5mC.txt — a simple text file with the following contents:
4m9e:B.5CM5: stacking-with-A.ARG443 is-WC-paired is-in-duplex [+]:GcG/cGC
4m9e:C.5CM5: other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG
- 4m9e-5mC.pdb — a corresponding PDB file, potentially multi-model, two as in this case. Moreover, the cluster of interacting residues (DNA nucleotides and protein amino acids) is oriented in the standard base reference frame of 5mC, allowing for easy comparison and direct overlap of multiple clusters.
In practice, SNAP needs to take care of many details for the automatic identification and annotation of 5mC-DNA-TF complexes directly from PDB entries. For example, 5mC in DNA is designated 5CM and the 5-methyl carbon atom is named C5A in the PDB (see the blogpost 5CM and 5MC, two forms of 5-methylcytosine in the PDB). Moreover, the --type=base option is employed to ensure that base atoms (regardless sugar-phosphate atoms) of 5mC are directly involved in interactions with amino acids.
It is also worth noting the combined use of DSSR for the generation of molecular images (rendered with PyMOL), as shown below. Here the DSSR options --block-file=fill-hbond (fill to fill base rings and hbond to draw hydrogen bonds) and --cartoon-block=sticks-label are used. The 3DNA DSSR/SNAP combo is a unique and powerful toolset for structural bioinformatics, as demonstrated in DNAproDB from the Rohs lab (see my blogpost SNAP and DSSR in DNAproDB). The JMB paper represents yet another example. I can only expect to see more combined DSSR/SNAP applications in the future.

I recently performed a quick survey of the cover images of the RNA journal in 2019. I was pleased to find that 9 out of the 12 cover images were provided by the Nucleic Acid Database where 3DNA/blockview and PyMOL were employed, as noted below:
The RNA backbone is displayed as a red ribbon; bases are shown as blocks with NDB coloring: A—red, C—yellow, G—green, U—cyan; geneticin ligands are shown in spacefill with element colors: C—white, N—blue, O—red. The image was generated using 3DNA/blocview and PyMol software.
Details of the 9 cover images are listed below:
- January 2019 Rhodobacter sphaeroides Argonaute with guide RNA/target DNA duplex containing noncanonical A-G pair (PDB code: 6d9k)
- April 2019 Group I self-splicing intron P4-P6 domain mutant U131A (PDB code: 6d8l)
- May 2019 Crystal structure of T. thermophilus 50S ribosomal protein L1 in complex with helices H76, H77, and H78 of 23S RNA (PDB code: 5npm)
- June 2019 Crystal structure of ykoY-mntP riboswitch chimera bound to cadmium (PDB code: 6cc3)
- July 2019 G96A mutant of the PRPP riboswitch from T. mathranii bound to ppGpp (PDB code: 6ck4)
- August 2019 Crystal structure of the metY SAM V riboswitch (PDB code: 6fz0)
- October 2019 Crystal structure of protease factor Xa bound to RNA aptamer 11F7t and rivaroxaban (PDB code: 5vof)
- November 2019 Drosophila melanogaster nucleosome remodeling complex (PDB code: 6f4g)
- December 2019 Crystal structure of the Homo Sapiens cytoplasmic ribosomal decoding site in complex with Geneticin (PDB code: 5xz1)
Here is the composite figure of the 9 cover images.

See also:

I’ve created a web API to DSSR and SNAP, and fiber models. The overall help message is available via http://api.x3dna.org. Individually, each program is accessed as below.
Usage with 'http' (HTTPie):
http -f http://api.x3dna.org/dssr [options] url=|model@
http http://api.x3dna.org/dssr/help -- display this help message
Options:
json=true-or-FALSE(default) [e.g., json=true # JSON output]
pair=true-or-FALSE(default) [e.g., pair=1 # base-pair only]
hbond=true-or-FALSE(default) [e.g., hbond=t # H-bonding info]
more=true-or-FALSE(default) [e.g., more=y # further details]
Required parameter:
url=URL-to-coordinate-file [e.g., url=https://files.rcsb.org/download/1ehz.pdb.gz]
model@coordinate-file [e.g., model@1ehz.cif]
# Only one must be specified. 'url' precedes 'model' when both are specified.
# The coordinate file must be in PDB or PDBx/mmCIF format, optionally gzipped.
Examples:
http -f http://api.x3dna.org/dssr url=https://files.rcsb.org/download/1ehz.pdb.gz
http -f http://api.x3dna.org/dssr model@1ehz.cif pair=1
# with 'curl'
curl http://api.x3dna.org/dssr -F 'url=https://files.rcsb.org/download/1ehz.pdb.gz'
curl http://api.x3dna.org/dssr -F 'model=@1msy.pdb' -F 'pair=1'
Note:
The web API has an upper limit on coordinate file size (gzipped): < 6 MB
Usage with 'http' (HTTPie):
http -f http://api.x3dna.org/snap [options] url=|model@
http http://api.x3dna.org/snap/help -- display this help message
Options:
json=true-or-FALSE(default) [e.g., json=true # JSON output]
hbond=true-or-FALSE(default) [e.g., hbond=t # H-bonding info]
Required parameter:
url=URL-to-coordinate-file [e.g., url=https://files.rcsb.org/download/1oct.pdb.gz]
model@coordinate-file [e.g., model@1oct.cif]
# Only one must be specified. 'url' precedes 'model' when both are specified.
# The coordinate file must be in PDB or PDBx/mmCIF format, optionally gzipped.
Examples:
http -f http://api.x3dna.org/snap url=https://files.rcsb.org/download/1oct.pdb.gz
http -f http://api.x3dna.org/snap model@1oct.cif json=1
# with 'curl'
curl http://api.x3dna.org/snap -F 'url=https://files.rcsb.org/download/1oct.pdb.gz'
curl http://api.x3dna.org/snap -F 'model=@1oct.cif' -F 'json=1'
Note:
The web API has an upper limit on coordinate file size (gzipped): < 6 MB
Usage with 'http' (HTTPie):
http http://api.x3dna.org/fiber/help # display this help message
http http://api.x3dna.org/fiber/list # show a list of available fiber models (56 in total)
http http://api.x3dna.org/fiber/str_id # build model 'str_id' in the range of [1, 56]
http http://api.x3dna.org/fiber/name # generate a model with common names as shown below:
A-DNA, B-dna, C_DNA, D-DNA, ZDNA, RNA, RNAduplex, PaulingTriplex, G4
Case does not matter, and the separator can be '-' or '_' or omitted.
So a-dna, A-dNA, a_DNA, or ADNA is valid for building an A-DNA model.
Options (via query strings, or form fields):
seq=base-sequence # A, C, G, T, U for generic model
repeat=number # number of repeats of the sequence
cif=1 # output file in mmCIF format
Examples with 'http' (HTTPie):
http http://api.x3dna.org/fiber/1 # model no. 1 (i.e., calf thymus A-DNA model)
http -f http://api.x3dna.org/fiber/1 seq=A3TTT repeat=2 # specific sequence, repeated twice
http http://api.x3dna.org/fiber/rna # single-stranded RNA model
http http://api.x3dna.org/fiber/rna-ds # double-stranded RNA model
http http://api.x3dna.org/fiber/pauling # the triplex model of Pauling & Corey
http http://api.x3dna.org/fiber/g4 # G-quadruplex model
# with 'curl'
curl http://api.x3dna.org/fiber/1
curl http://api.x3dna.org/fiber/1 -d 'seq=A3TTT' -d 'repeat=2'
curl http://api.x3dna.org/fiber/rna
curl http://api.x3dna.org/fiber/rna-ds
curl http://api.x3dna.org/fiber/pauling
curl http://api.x3dna.org/fiber/g4
Note:
The web API has two upper limits: repeats < 1,000, and nucleotides < 10,000.