Cover images provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
See the 2020 paper titled "DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL" in Nucleic Acids Research and the corresponding Supplemental PDF for details. Many thanks to Drs. Wilma Olson and Cathy Lawson for their help in the preparation of the illustrations.
Details on how to reproduce the cover images are available on the 3DNA Forum.

Structure of the human minor spliceosome pre-B complex (PDB id: 8Y7E; Bai R, Yuan M, Zhang P, Luo T, Shi Y, Wan R. 2024. Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome. Science 383: 1245–1252). The protein–RNA assembly reveals the mechanisms of recognition and recruitment of several small nuclear ribonucleoproteins (snRNPs) involved in the splicing of U12-type introns. The pre-mRNA is depicted by a red ribbon, and the U12 small nuclear RNA (snRNA) by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are shown as gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Human tRNA splicing endonuclease (TSEN) complex bound to pre-tRNAArg (PDB id: 7UXA; Hayne CK, Butay KJ, Stewart ZD, Krahn JM, Perera L, Williams JG, Petrovitch RM, Deterding LJ, Matera AG, Borgnia MJ, Stanley RE. 2023. Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex. Nat Struct Mol Biol 30: 824–833). Cryo-EM structure of the TSEN protein assembly with pre-tRNAArg provides insights into the recognition and splicing of an intron that must be removed from the pre-tRNA before translation. The pre-tRNAArg is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the TSEN subunits are shown as gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Systemic RNA interference defective protein 1 (SID1) in complex with dsRNA (PDB id: 8XC1; Wang R, Cong Y, Qian D, Yan C, Gong D. 2024. Structural basis for double-stranded RNA recognition by SID1. Nucleic Acids Res 52: 6718–6727). The cryo-EM structure provides a major step towards understanding the mechanism of dsRNA recognition by SID1, involving extensive interactions between basic amino-acid residues and the sugar-phosphate backbone. The dsRNA chains are depicted by red, green, blue, and yellow ribbons, with bases and Watson-Crick base pairs represented as color-coded blocks and minor-groove edges colored white: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; SID1 is shown by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Complex of arginyl-tRNA-protein transferase 1 (ATE1) with tRNAArg and a short peptide substrate (PDB id: 8UAU; Lan X, Huang W, Kim SB, Fu D, Abeywansha T, Lou J, Balamurugan U, Kwon YT, Ji CH, Taylor DJ, Zhang Y. 2024. Oligomerization and a distinct tRNA-binding loop are important regulators of human arginyl-transferase function. Nat Commun 15: 6350). The ATE1 homodimer dissociates upon binding the peptide and forms a loop that wraps around tRNAArg. The tRNAArg is depicted by a red ribbon, with bases and Watson–Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; ATE1 is shown by a gold ribbon and the peptide by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of endoribonuclease P (RNase P) in complex with pre-tRNAHis-Ser (PDB id: 8CBK; Meynier V, Hardwick SW, Catala M, Roske JJ, Oerum S, Chirgadze DY, Barraud P, Yue WW, Luisi BF, Tisné C. 2024. Structural basis for human mitochondrial tRNA maturation. Nat Commun 15: 4683). The structure reveals the first step of human mitochondrial tRNA maturation by RNase P, processing the 5′-leader of pre-tRNA. The RNA is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the protein assembly is shown by the gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of a group II intron ribonucleoprotein in the pre-ligation state (PDB id: 8T2R; Xu L, Liu T, Chung K, Pyle AM. 2023. Structural insights into intron catalysis and dynamics during splicing. Nature 624: 682–688). The pre-ligation complex of the Agathobacter rectalis group II intron reverse transcriptase/maturase with intron and 5′-exon RNAs makes it possible to construct a picture of the splicing active site. The intron is depicted by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the 5′-exon is shown by white spheres and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Complex of terminal uridylyltransferase 7 (TUT7) with pre-miRNA and Lin28A (PDB id: 8OPT; Yi G, Ye M, Carrique L, El-Sagheer A, Brown T, Norbury CJ, Zhang P, Gilbert RJ. 2024. Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs. Nat Struct Mol Biol 31: 1426–1438). The RNA-binding pluripotency factor LIN28A invades and melts the RNA and affects the mechanism of action of the TUT7 enzyme. The RNA backbone is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; TUT7 is represented by a gold ribbon and LIN28A by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Cryo-EM structure of the pre-B complex (PDB id: 8QP8; Zhang Z, Kumar V, Dybkov O, Will CL, Zhong J, Ludwig SE, Urlaub H, Kastner B, Stark H, Lührmann R. 2024. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 630: 1012–1019). The pre-B complex is thought to be critical in the regulation of splicing reactions. Its structure suggests how the cross-exon and cross-intron spliceosome assembly pathways converge. The U4, U5, and U6 snRNA backbones are depicted respectively by blue, green, and red ribbons, with bases and Watson-Crick base pairs shown as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are represented by gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the Hendra henipavirus (HeV) nucleoprotein (N) protein-RNA double-ring assembly (PDB id: 8C4H; Passchier TC, White JB, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN. 2024. The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Sci Rep 14: 14099). The HeV N protein adopts a bi-lobed fold, where the N- and C-terminal globular domains are bisected by an RNA binding cleft. Neighboring N proteins assemble laterally and completely encapsidate the viral genomic and antigenomic RNAs. The two RNAs are depicted by green and red ribbons. The U bases of the poly(U) model are shown as cyan blocks. Proteins are represented as semitransparent gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the helicase and C-terminal domains of Dicer-related helicase-1 (DRH-1) bound to dsRNA (PDB id: 8T5S; Consalvo CD, Aderounmu AM, Donelick HM, Aruscavage PJ, Eckert DM, Shen PS, Bass BL. 2024. Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA. eLife 13: RP93979. Cryo-EM structures of Dicer-1 in complex with DRH-1, RNAi deficient-4 (RDE-4), and dsRNA provide mechanistic insights into how these three proteins cooperate in antiviral defense. The dsRNA backbone is depicted by green and red ribbons. The U-A pairs of the poly(A)·poly(U) model are shown as long rectangular cyan blocks, with minor-groove edges colored white. The ADP ligand is represented by a red block and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Moreover, the following 30 [12(2021) + 12(2022) + 6(2023)] cover images of the RNA Journal were generated by the NAKB (nakb.org).
Cover image provided by the Nucleic Acid Database (NDB)/Nucleic Acid Knowledgebase (NAKB; nakb.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

With great pleasure, I read the following annoancement from Rajendra Kumar on the 3DNA Forum:
Re: do_x3dna: a tool to analyze DNA/RNA in molecular dynamics trajectories
« Reply #1 on: Today at 10:53:31 AM »
Hello,
I have now made a new website for do_x3dna
(http://rjdkmr.github.io/do_x3dna). This website contains detailed
documentation for do_x3dna program and Python APIs.
Documentation for Python API is now available
(http://rjdkmr.github.io/do_x3dna/apidoc.html).
Few tutorials about the Python APIs are also now available
(http://rjdkmr.github.io/do_x3dna/tutorial.html).
Thanks.
With best regards,
Rajendra
Browsing through the do_x3dna website, I am impressed by the extensive documentation and tutorial. Clearly, do_x3dna has pushed the boundaries (in applicability and documentation) of the x3dna_ensemble Ruby script distributed with 3DNA v2.1.
As noted in GitHub page, do_x3dna has been developed to analyze fluctuations in DNA or RNA structures in molecular dynamics (MD) trajectories. It can be used for GROMACS MD trajectories, as well as those from NAMD and AMBER. It leaves no doubt that do_x3dna will boost 3DNA’s applications in the increasingly active field of DNA/RNA MD simulations.

From early on, 3DNA and DSSR have native support of modified nucleotides. The currently distributed baselist.dat file with 3DNA contains over 700 entries. As of v1.1.4-2014aug09, a new section has been added to DSSR to list explicitly the modified nucleotides in an analyzed structure.
Using the 76-nucleotide long yeast phenylalanine tRNA (1ehz) as an example, the pertinent section in DSSR output is as below.
List of 11 types of 14 modified nucleotides
nt count list
1 1MA-a 1 A.1MA58
2 2MG-g 1 A.2MG10
3 5MC-c 2 A.5MC40,A.5MC49
4 5MU-t 1 A.5MU54
5 7MG-g 1 A.7MG46
6 H2U-u 2 A.H2U16,A.H2U17
7 M2G-g 1 A.M2G26
8 OMC-c 1 A.OMC32
9 OMG-g 1 A.OMG34
10 PSU-P 2 A.PSU39,A.PSU55
11 YYG-g 1 A.YYG37
So 1ehz has 14 modified nucleotides of 11 different type, as listed in the following rows after the header line. The meaning of each column should be obvious. For example, the third row means that 5MC (5-methylcytidine, abbreviated as 'c' in 1-letter code) occurs twice, identified as A.5MC40 and A.5MC49, respectively.
With the 3-letter id, one can search the RCSB ligand database for more information about a specified modified nucleotide. The URL would be like this, using pseudouridine (PSU) as an example, https://www.rcsb.org/ligand/PSU.
It is hoped that the newly added section, put at the very top of DSSR output, will draw more attention to modified nucleotides.

From v1.1.3-2014jun18, DSSR has an additional output of RNA secondary structures in BPSEQ format. A sample file for PDB entry 1msy is shown below.
![1msy [GUAA tetra loop] in 3d and 2d representations 1msy [GUAA tetra loop] in 3d and 2d representations](http://forum.x3dna.org/images/1msy-3d-2d.png)
Filename: dssr-2ndstrs.bpseq
Organism: DSSR-derived secondary structure [1msy]
Accession Number: DSSR v1.1.4-2014aug09 (xiangjun@x3dna.org)
Citation: Please cite 3DNA/DSSR (see http://home.x3dna.org)
1 U 0 # name=A.U2647
2 G 26 # name=A.G2648, pairedNt=A.U2672
3 C 25 # name=A.C2649, pairedNt=A.G2671
4 U 24 # name=A.U2650, pairedNt=A.A2670
5 C 23 # name=A.C2651, pairedNt=A.G2669
6 C 22 # name=A.C2652, pairedNt=A.G2668
7 U 0 # name=A.U2653
8 A 0 # name=A.A2654
9 G 0 # name=A.G2655
10 U 0 # name=A.U2656
11 A 0 # name=A.A2657
12 C 17 # name=A.C2658, pairedNt=A.G2663
13 G 0 # name=A.G2659
14 U 0 # name=A.U2660
15 A 0 # name=A.A2661
16 A 0 # name=A.A2662
17 G 12 # name=A.G2663, pairedNt=A.C2658
18 G 0 # name=A.G2664
19 A 0 # name=A.A2665
20 C 0 # name=A.C2666
21 C 0 # name=A.C2667
22 G 6 # name=A.G2668, pairedNt=A.C2652
23 G 5 # name=A.G2669, pairedNt=A.C2651
24 A 4 # name=A.A2670, pairedNt=A.U2650
25 G 3 # name=A.G2671, pairedNt=A.C2649
26 U 2 # name=A.U2672, pairedNt=A.G2648
27 G 0 # name=A.G2673
Based on online sources, BPSEQ has originated from the Comparative RNA Web site developed by the Gutell lab. CRW files contain four header lines, describing the file name, organism, accession number, and a general remark. Thereafter, there is one line per base in the molecule, listing the position of the base (starting from 1), the one-letter base name (A,C,G,U etc), and the position number of the base to which it is paired. If the base is unpaired, zero (0) is put in the third column. In the above sample BPSEQ file derived from DSSR, detailed information about the base and its paired base (if any) comes after the # symbol.
Compared to dot-bracket notation (dbn) and connect-table (.ct) format, BPSEQ is simpler but less expressive. Nevertheless, the format is well-supported in bioinformatic tools on RNA secondary structures. It only seems fitting that DSSR now produces secondary structures in .bpseq (with default file name dssr-2ndstrs.bpseq), in addition to .dbn and .ct. Technically, adding the BPSEQ output to DSSR is trivial given the infrastructure already in place.

From early on, DSSR-derived RNA secondary structures in dot-bracket notation (dbn) have taken pseudoknots into consideration. Nevertheless, in DSSR releases prior to v1.1.3-2014jun18, the dbn output had been simplified to the first level only, with matched []s, even for RNA structures with high-order pseudoknots. RNA pseudoknot is a (relatively) complicated issue, and I’d planned to put off the topic until DSSR is well-established.
In early May, I noticed the Antczak et al. article RNApdbee—a webserver to derive secondary structures from pdb files of knotted and unknotted RNAs. I was delighted to read the following citation:
In order to facilitate a more comprehensive study, the webserver integrates the functionality of RNAView, MC-Annotate and 3DNA/DSSR, being the most common tools used for automated identification and classification of RNA base pairs.
Even before any paper on DSSR has been published, the software has already be ranked in the top three for the identification and classification of RNA base pairs! Well familiar with RNAView and MC-Annotate, I am glad to see DSSR is now listed on a par with them. Note that DSSR has far more functionality than just identifying and classifying RNA base pairs.
Further down the RNApdbee paper, especially in Figure 2, I found the following remarks regarding DSSR’s capability on RNA structures with high-order pseudoknot.
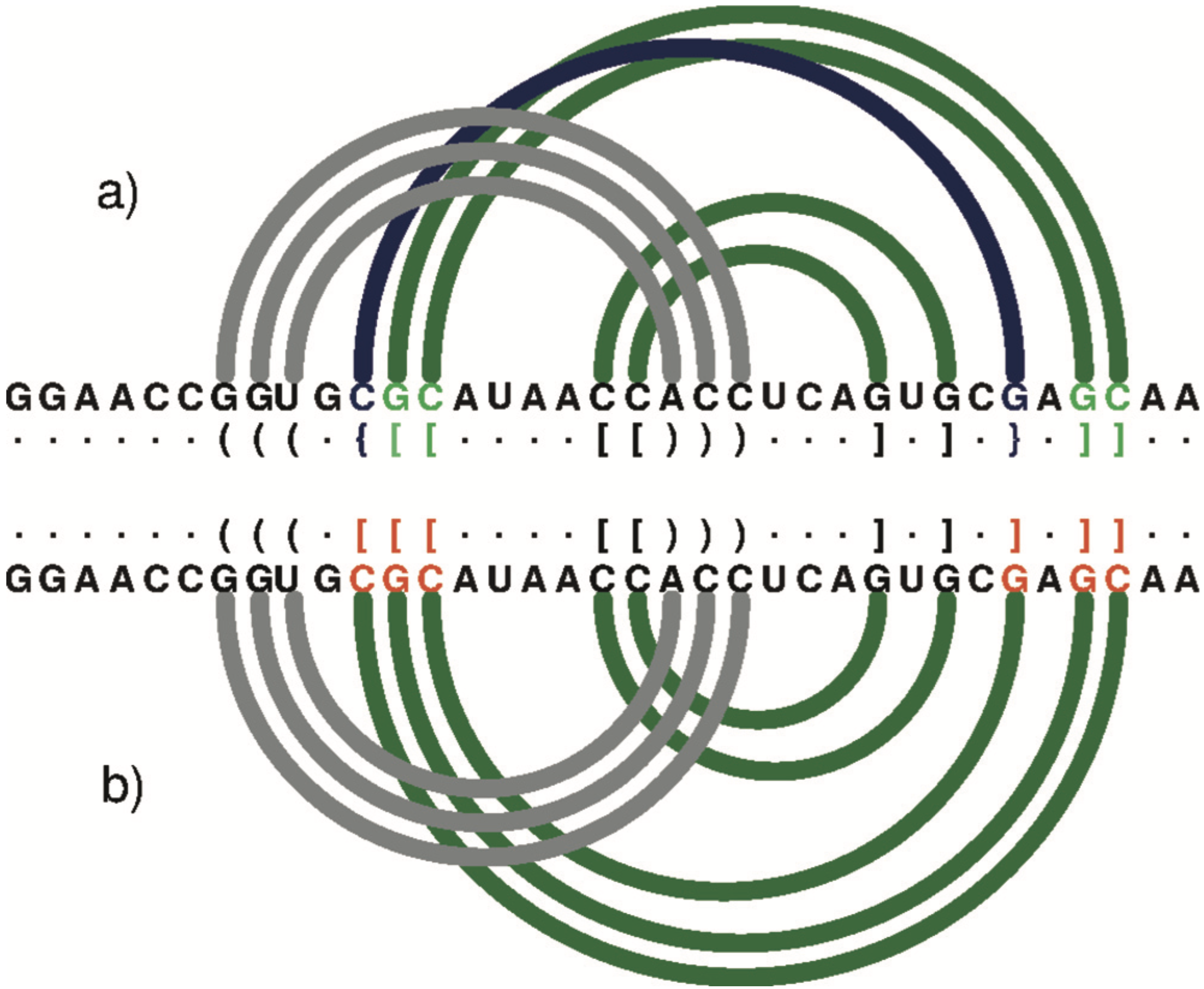
An arc diagram to represent the secondary structure of 1DDY (chain A) generated by R-CHIE upon the dot-bracket notation. Arcs of the same colour define a paired region. Crossing arcs reflect a conflict observed between the corresponding regions. (a) RNApdbee recognizes pseudoknots of the first (dark green) and second (navy blue) order. (b) 3DNA/DSSR improperly classifies base pairs (within residues in red) and the structure is recognized as the first-order pseudoknot.
The above citation and the question Higher-order pseudoknots in DP output (from Jan Hajic, Charles University in Prague) on the 3DNA Forum prompted me to further refine DSSR’s algorithm for deriving secondary structures of RNA with high-order pseudoknots. The DSSR v1.1.3-2014jun18 release made this revised functionality explicit. For the above cited PDB entry 1ddy, the relevant output of running DSSR on it would be:
Running command: "x3dna-dssr -i=1ddy.pdb"
****************************************************************************
This structure contains 2-order pseudoknot(s)
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>1ddy nts=140 [whole]
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA&GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA&GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA&GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((.{[[....[[)))...].].}.]]..&......(((.{[[....[[)))...].].}.]]..&......(((.{[[....[[)))...].].}.]]..&......(((.{[[....[[)))...].].}.]]..
>1ddy-A #1 nts=35 [chain] RNA
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((.{[[....[[)))...].].}.]]..
>1ddy-C #2 nts=35 [chain] RNA
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((.{[[....[[)))...].].}.]]..
>1ddy-E #3 nts=35 [chain] RNA
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((.{[[....[[)))...].].}.]]..
>1ddy-G #4 nts=35 [chain] RNA
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((.{[[....[[)))...].].}.]]..
Note that the whole 1ddy entry contains four RNA chains (A, C, E, and G), and DSSR can handle each properly. So at least from DSSR v1.1.3-2014jun18, the following statement is no longer valid:
3DNA/DSSR improperly classifies base pairs (within residues in red) and the structure is recognized as the first-order pseudoknot.
A closely related issue is knot removal, a topic nicely summarized by Smit et al. in their publication From knotted to nested RNA structures: A variety of computational methods for pseudoknot removal. While not explicitly documented, the --nested (abbreviated to --nest) option has been available since DSSR v1.1.3-2014jun18. This option was first mentioned in the release note of DSSR v1.1.4-2014aug09. Again, using PDB entry 1ddy as an example, the relevant output of running DSSR with option --nested is as follows:
Running command: "x3dna-dssr -i=1ddy.pdb --nested"
****************************************************************************
This structure contains 2-order pseudoknot(s)
o You've chosen to remove pseudo-knots, leaving only nested pairs
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>1ddy nts=140 [whole]
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA&GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA&GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA&GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((..........))).............&......(((..........))).............&......(((..........))).............&......(((..........))).............
>1ddy-A #1 nts=35 [chain] RNA
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((..........))).............
>1ddy-C #2 nts=35 [chain] RNA
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((..........))).............
>1ddy-E #3 nts=35 [chain] RNA
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((..........))).............
>1ddy-G #4 nts=35 [chain] RNA
GGAACCGGUGCGCAUAACCACCUCAGUGCGAGCAA
......(((..........))).............

H-bonding interactions are crucial for defining RNA secondary and tertiary structures. DSSR/3DNA contains a geometrically based algorithm for identifying H-bonds in nucleic-acid or protein structures given in .pdb or .cif format. Over the years, the method has been continuously refined, and it has served its purpose quite well. As of v1.1.1-2014apr11, this functionality is directly available from DSSR thorough the --get-hbonds option.
The output for 1msy, which contains a GUAA tetraloop mutant of Sarcin/Ricin domain from E. Coli 23 S rRNA, is listed below. The first line gives the header (# H-bonds in '1msy.pdb' identified by DSSR ...). The second line provides the total number of H-bonds (40) identified in the structure. Afterwards, each line consists of 8 space-delimited columns used to characterize a specific H-bond. Using the first one (#1) as an example, the meaning of each of the 8 columns is:
- The serial number (15), as denoted in the .pdb or .cif file, of the first atom of the H-bond.
- The serial number (578) of the second H-bond atom.
- The H-bond index (#1), from 1 to the total number of H-bonds.
- A one-letter symbol showing the atom-pair type (p) of the H-bond. It is ‘p’ for a donor-acceptor atom pair; ‘o’ for a donor/acceptor (such as the 2′-hydorxyl oxygen) with any other atom; ‘x’ for a donor-donor or acceptor-acceptor pair (as in #17); ‘?’ if the donor/acceptor status is unknown for any H-bond atom.
- Distance in Å between donor/acceptor atoms (2.768).
- Elemental symbols of the two atoms involved in the H-bond (O/N).
- Identifier of the first H-bond atom (O4@A.U2647).
- Identifier of the second H-bond atom (N1@A.G2673).
Command: x3dna-dssr -i=1msy.pdb --get-hbonds –o=1msy-hbonds.txt
# H-bonds in '1msy.pdb' identified by 3DNA version 3 (xiangjun@x3dna.org)
40
15 578 #1 p 2.768 O:N O4@A.U2647 N1@A.G2673
35 555 #2 p 2.776 O:N O6@A.G2648 N3@A.U2672
36 554 #3 p 2.826 N:O N1@A.G2648 O2@A.U2672
55 537 #4 p 2.965 O:N O2@A.C2649 N2@A.G2671
56 535 #5 p 2.836 N:N N3@A.C2649 N1@A.G2671
58 534 #6 p 2.769 N:O N4@A.C2649 O6@A.G2671
76 513 #7 p 2.806 N:N N3@A.U2650 N1@A.A2670
78 512 #8 p 3.129 O:N O4@A.U2650 N6@A.A2670
95 492 #9 p 2.703 O:N O2@A.C2651 N2@A.G2669
96 490 #10 p 2.853 N:N N3@A.C2651 N1@A.G2669
98 489 #11 p 2.987 N:O N4@A.C2651 O6@A.G2669
115 466 #12 p 2.817 O:N O2@A.C2652 N2@A.G2668
116 464 #13 p 2.907 N:N N3@A.C2652 N1@A.G2668
118 463 #14 p 2.897 N:O N4@A.C2652 O6@A.G2668
123 151 #15 o 2.622 O:O OP2@A.U2653 O2'@A.A2654
135 443 #16 p 2.898 O:N O2@A.U2653 N4@A.C2667
147 192 #17 x 3.054 O:O O4'@A.A2654 O4'@A.U2656
158 408 #18 p 2.960 N:O N6@A.A2654 OP2@A.C2666
173 188 #19 o 2.923 O:O O2'@A.G2655 OP2@A.U2656
173 378 #20 o 3.093 O:O O2'@A.G2655 O6@A.G2664
173 379 #21 o 3.343 O:N O2'@A.G2655 N1@A.G2664
181 386 #22 p 2.768 N:O N1@A.G2655 OP2@A.A2665
183 203 #23 p 2.754 N:O N2@A.G2655 O4@A.U2656
183 387 #24 p 2.887 N:O N2@A.G2655 O5'@A.A2665
188 379 #25 p 3.044 O:N OP2@A.U2656 N1@A.G2664
188 381 #26 p 2.944 O:N OP2@A.U2656 N2@A.G2664
200 401 #27 p 3.122 O:N O2@A.U2656 N6@A.A2665
201 398 #28 p 2.759 N:N N3@A.U2656 N7@A.A2665
220 381 #29 p 3.035 N:N N7@A.A2657 N2@A.G2664
223 371 #30 o 2.963 N:O N6@A.A2657 O2'@A.G2664
223 382 #31 p 3.039 N:N N6@A.A2657 N3@A.G2664
242 358 #32 p 2.821 O:N O2@A.C2658 N2@A.G2663
243 356 #33 p 2.890 N:N N3@A.C2658 N1@A.G2663
245 355 #34 p 2.887 N:O N4@A.C2658 O6@A.G2663
258 305 #35 o 2.604 O:N O2'@A.G2659 N7@A.A2661
258 308 #36 o 3.264 O:N O2'@A.G2659 N6@A.A2661
268 315 #37 p 2.973 N:O N2@A.G2659 OP2@A.A2662
268 327 #38 p 2.864 N:N N2@A.G2659 N7@A.A2662
371 390 #39 o 2.751 O:O O2'@A.G2664 O4'@A.A2665
550 566 #40 o 3.372 O:O O2'@A.U2672 O4'@A.G2673
In its default settings, DSSR detects 117 H-bonds for 1ehz (yeast phenylalanine tRNA), and 5,809 for 1jj2 (the H. marismortui large ribosomal subunit). Note that the program can identify H-bonds not only in RNA and DNA, but also in proteins, or their complexes. By default, however, DSSR only reports H-bonds within nucleic acids. As shown above, it is trivial to run DSSR with the --get-hbonds option to get all H-bonds in a given structure, and the plain text output is straightforward to work on.
While there exist dedicated tools for finding H-bonds, such as HBPLUS or HBexplore, DSSR may well be sufficient to fulfill most practical needs. If you notice any weird behaviors with this H-bond finding functionality, please let me know. I strive to address reported issues promptly, to the extent practical. At the very least, I should be able to explain why the program is working the way it does.

From the very first release up until recently, the DSSR distribution had included two executables for Windows: one version was compiled on MinGW/MSYS, and the other on Cygwin. The executables are supposed to be run under the corresponding shells of the two environments respectively.
Since DSSR is a simple self-contained command-line tool, the MinGW/MSYS version also works directly under the Command Prompt of native Windows. So Windows users had the following three options to use DSSR:
- Download the MinGW/MSYS version to run it under the Command Prompt of native Windows. No need to install MinGW/MSYS.
- Download the MinGW/MSYS version to run it under the MinGW/MSYS environment, which must be installed separately.
- Download the Cygwin version to run it under the Cygwin environment, which must be installed separately.
Over times, I have observed some confusions among DSSR users as to which of the two executables to use on Windows. Luckily, I noticed by chance recently that the DSSR executable compiled under MinGW/MSYS runs just fine in the Cygwin shell. So as of v1.1.0-2014apr09, the DSSR distribution contains only one executable for Windows: compiled under MinGW/MSYS on 32-bit Windows XP, the same DSSR executable runs under the Command Prompt of native Windows, MinGW/MSYS, and Cygwin, either on a 32-bit or 64-bit Windows (XP, Vista, 7 or 8) machine.
A size fits all: I no longer need to provide two compiled versions of DSSR for Windows, and users have just one executable to download (no more space for confusions).
Note added on 2024-11-25: DSSR is distributed by the CTV (Columbia Technology Ventures). See https://home.x3dna.org

In addition to VARNA, the draw program in the RNAstructure package from the Mathews Laboratory can also be used to depict DSSR-derived RNA secondary structures in connect table (.ct) format. The draw program produces images in PostScript (or svg) format, in different styles from those generated by VARNA. Given below are a couple of examples on how to connect DSSR with draw.
The secondary structure of the PDB entry 1msy in DSSR-derived .ct file is as below:
27 DSSR-derived secondary structure in '1msy'
1 U 0 2 0 2647
2 G 1 3 26 2648
3 C 2 4 25 2649
4 U 3 5 24 2650
5 C 4 6 23 2651
6 C 5 7 22 2652
7 U 6 8 0 2653
8 A 7 9 0 2654
9 G 8 10 0 2655
10 U 9 11 0 2656
11 A 10 12 0 2657
12 C 11 13 17 2658
13 G 12 14 0 2659
14 U 13 15 0 2660
15 A 14 16 0 2661
16 A 15 17 0 2662
17 G 16 18 12 2663
18 G 17 19 0 2664
19 A 18 20 0 2665
20 C 19 21 0 2666
21 C 20 22 0 2667
22 G 21 23 6 2668
23 G 22 24 5 2669
24 A 23 25 4 2670
25 G 24 26 3 2671
26 U 25 27 2 2672
27 G 26 0 0 2673
Let the DSSR-derived .ct file for 1msy be named 1msy.ct, the following two draw-command runs will produce the secondary structure in PostScript (1msy.eps) and svg (1msy.svg) respectively.
draw 1msy.ct 1msy.eps
draw 1msy.ct 1msy.svg --svg -n 1
![1msy [GUAA tetra loop] 2nd structure produced with the RNAstructure 'draw' program 1msy [GUAA tetra loop] 2nd structure produced with the RNAstructure 'draw' program](http://forum.x3dna.org/images/1msy.svg)
The PDB entry 1ehz (yeast phenylalanine tRNA) has a pseudo knot, so the draw program will create a ‘circularized’ structure as shown below:
![1ehz [yeast phenylalanine tRNA] 2nd structure produced with the RNAstructure 'draw' program 1ehz [yeast phenylalanine tRNA] 2nd structure produced with the RNAstructure 'draw' program](http://forum.x3dna.org/images/1ehz.svg)
Note the following two caveats:

As of v1.0.3-2014mar09, DSSR has a decent user manual in PDF! Currently of 45 pages long, the DSSR manual contains everything a typical user needs to know to get started using the program effectively. The contents the manual are listed below.
Table of Contents
List of Figures
Introduction
Download and installation
Usages
Command-line help
Default run on PDB entry 1msy – detailed explanations
Summary section
List of base pairs
List of multiplets
List of helices
List of stems
List of lone canonical pairs
List of various loops
List of single-stranded fragments
Secondary structure in dot-bracket notation
List of backbone torsion angles and suite names
Default run on PDB entry 1ehz (tRNAPhe) – summary notes
Brief summary
Specific features
Default run on PDB entry 1jj2 – four auto-checked motifs
Kissing loops
A-minor (types I and II) motifs
Ribose zippers
Kink turns
The --more option
Extra parameters for base pairs
Extra parameters for helices/stems
The –-non-pair option
The –-u-turn option
The --po4 option
The –-long-idstr option
Frequently asked questions
How to cite DSSR?
Does DSSR work for DNA?
Does DSSR detect RNA tertiary interactions?
Revision history
Acknowledgements
References
With the User Manual available, I feel confident to claim that DSSR is now mature, stable, ready for real world applications. While only time would tell, I have no doubt that DSSR will become an essential tool in RNA structural bioinformatics.

From early on, DSSR-derived nucleic acid secondary structures have been written in the compact dot-bracket notation (.dbn) with pseudo-knot information. To better connect DSSR to the 2D world, I recently looked into the connect (.ct) format, which was first introduced by Zuker’s mfold program. Over time, the .ct format has become one of the most commonly used RNA secondary structure formats, and it is more expressive than the .dbn format (see below).
As of v1.0, for each analyzed structure, DSSR produces two secondary structure files with default names dssr-2ndstrs.dbn and dssr-2ndstrs.ct, in .dbn and .ct formats, respectively. Using the 27-nucleotides (nt) RNA fragment 1msy as an example, the DSSR-derived secondary structure in .dbn and .ct formats are shown below:
![1msy [GUAA tetra loop] in 3d and 2d representations 1msy [GUAA tetra loop] in 3d and 2d representations](http://forum.x3dna.org/images/1msy-3d-2d.png)
In dot-bracket notation (.dbn) [dssr-2ndstrs.dbn]
------------------------------------------------------
>1msy nts=27 DSSR-derived secondary structure
UGCUCCUAGUACGUAAGGACCGGAGUG
.(((((.....(....)....))))).
------------------------------------------------------
In connect format (.ct) [dssr-2ndstrs.ct]
------------------------------------------------------
27 DSSR-derived secondary structure in '1msy'
1 U 0 2 0 2647 # name=A.U2647
2 G 1 3 26 2648 # name=A.G2648, pairedNt=A.U2672
3 C 2 4 25 2649 # name=A.C2649, pairedNt=A.G2671
4 U 3 5 24 2650 # name=A.U2650, pairedNt=A.A2670
5 C 4 6 23 2651 # name=A.C2651, pairedNt=A.G2669
6 C 5 7 22 2652 # name=A.C2652, pairedNt=A.G2668
7 U 6 8 0 2653 # name=A.U2653
8 A 7 9 0 2654 # name=A.A2654
9 G 8 10 0 2655 # name=A.G2655
10 U 9 11 0 2656 # name=A.U2656
11 A 10 12 0 2657 # name=A.A2657
12 C 11 13 17 2658 # name=A.C2658, pairedNt=A.G2663
13 G 12 14 0 2659 # name=A.G2659
14 U 13 15 0 2660 # name=A.U2660
15 A 14 16 0 2661 # name=A.A2661
16 A 15 17 0 2662 # name=A.A2662
17 G 16 18 12 2663 # name=A.G2663, pairedNt=A.C2658
18 G 17 19 0 2664 # name=A.G2664
19 A 18 20 0 2665 # name=A.A2665
20 C 19 21 0 2666 # name=A.C2666
21 C 20 22 0 2667 # name=A.C2667
22 G 21 23 6 2668 # name=A.G2668, pairedNt=A.C2652
23 G 22 24 5 2669 # name=A.G2669, pairedNt=A.C2651
24 A 23 25 4 2670 # name=A.A2670, pairedNt=A.U2650
25 G 24 26 3 2671 # name=A.G2671, pairedNt=A.C2649
26 U 25 27 2 2672 # name=A.U2672, pairedNt=A.G2648
27 G 26 0 0 2673 # name=A.G2673
------------------------------------------------------
Presumably, the .ct format is very simple, and examining a sample file as shown above would give one a pretty good sense of what each column is about. While there exist many oversimplified descriptions of the .ct format on the web, the most detailed and accurate explanation is from the mfold manual:
The ``ct’‘ file (connect table) contains the sequence and base pair information, and is meant to be an input file for a structure drawing program. In addition to containing base pair information, it also lists the 5′ and 3′ neighbor of each base, allowing for the representation of circular RNA or multiple molecules. The ct file also lists the historical base numbering in the original sequence, as bases and base pairs are numbered according from 1 to the size of the folded segment. A portion of a ct file is displayed in Figure 12.
Figure 12: The ct file for the second and final folding of S. cerevisiae Phe-tRNA at 37°, with default parameters. The first record displays the fragment size (76), ΔG and sequence name. The ith subsequent record contains, in order, i, ri, the index of the 5′-connecting base, the index of the 3′-connecting base, the index of the paired base and the historical numbering of the ith base in the original sequence. The 5′, 3′ and base pair indices are 0 when there is no connection or base pair.
Specifically, the 3rd, 4th, and 6th columns in the .ct format convey specific information; by design, they are not redundant to information contained in the 1st column. Note that in the above ‘1msy’ example, the 6th column gives the nt sequence numbers (as in the PDB datafile) instead of the serial numbers (as in the 1st column). The DSSR produced .ct files also contain extra information after ‘#’, in the comma separated key=value format.
As an example of the usefulness of the 3rd and 4th columns, have a look of the DSSR-derived .ct file for the Dickerson DNA dodecamer duplex with sequence CGCGAATTCGCG:
24 DSSR-derived secondary structure in '355d'
1 C 0 2 24 1 # name=A.DC1, pairedNt=B.DG24
2 G 1 3 23 2 # name=A.DG2, pairedNt=B.DC23
3 C 2 4 22 3 # name=A.DC3, pairedNt=B.DG22
4 G 3 5 21 4 # name=A.DG4, pairedNt=B.DC21
5 A 4 6 20 5 # name=A.DA5, pairedNt=B.DT20
6 A 5 7 19 6 # name=A.DA6, pairedNt=B.DT19
7 T 6 8 18 7 # name=A.DT7, pairedNt=B.DA18
8 T 7 9 17 8 # name=A.DT8, pairedNt=B.DA17
9 C 8 10 16 9 # name=A.DC9, pairedNt=B.DG16
10 G 9 11 15 10 # name=A.DG10, pairedNt=B.DC15
11 C 10 12 14 11 # name=A.DC11, pairedNt=B.DG14
12 G 11 0 13 12 # name=A.DG12, pairedNt=B.DC13
13 C 0 14 12 13 # name=B.DC13, pairedNt=A.DG12
14 G 13 15 11 14 # name=B.DG14, pairedNt=A.DC11
15 C 14 16 10 15 # name=B.DC15, pairedNt=A.DG10
16 G 15 17 9 16 # name=B.DG16, pairedNt=A.DC9
17 A 16 18 8 17 # name=B.DA17, pairedNt=A.DT8
18 A 17 19 7 18 # name=B.DA18, pairedNt=A.DT7
19 T 18 20 6 19 # name=B.DT19, pairedNt=A.DA6
20 T 19 21 5 20 # name=B.DT20, pairedNt=A.DA5
21 C 20 22 4 21 # name=B.DC21, pairedNt=A.DG4
22 G 21 23 3 22 # name=B.DG22, pairedNt=A.DC3
23 C 22 24 2 23 # name=B.DC23, pairedNt=A.DG2
24 G 23 0 1 24 # name=B.DG24, pairedNt=A.DC1
Note the 0 at the 4th column for A.DG12 which is at the 3′ end of chain A, and the 0 at 3rd column for B.DC13 which is at the 5′ end of chain B.













![1msy [GUAA tetra loop] in 3d and 2d representations 1msy [GUAA tetra loop] in 3d and 2d representations](http://forum.x3dna.org/images/1msy-3d-2d.png)

![1msy [GUAA tetra loop] 2nd structure produced with the RNAstructure 'draw' program 1msy [GUAA tetra loop] 2nd structure produced with the RNAstructure 'draw' program](http://forum.x3dna.org/images/1msy.svg)
![1ehz [yeast phenylalanine tRNA] 2nd structure produced with the RNAstructure 'draw' program 1ehz [yeast phenylalanine tRNA] 2nd structure produced with the RNAstructure 'draw' program](http://forum.x3dna.org/images/1ehz.svg)